During cell division, the genome is fully duplicated and then segregated to two daughter cells. In some tissues, however, cells repeatedly duplicate their genome and grow without dividing. This process results in large cells with many copies of their genome, a state known as polyploidy. In polyploid cells of the fruit fly Drosophila, DNA replication is not complete, which results in a relatively lower DNA copy number of specific genomic loci. In PNAS, Sher et al. show that in two different polyploid cell types of mice the entire genome is fully duplicated, unlike in Drosophila, and that this organismal difference may be explained by different levels of expression of the genes required for DNA replication (1). This report from Sher et al. reveals how these variant polyploid cell cycles can differ among organisms and tissues, but also uncovers many key similarities between flies and mammals that provide clues to the regulation of these enigmatic cell cycles.
Developmental Variations on the Cell Cycle Theme: Endocycles and Endomitosis
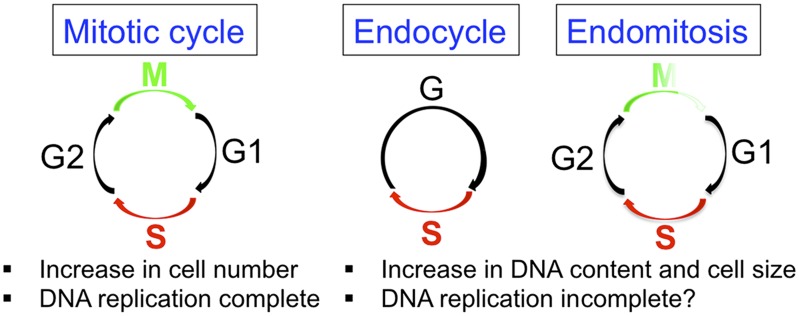
A common type of polyploid cycle is called the endocycle, which is comprised of alternating DNA synthesis (S) phases and gap (G) phases without chromosome segregation during a mitotic (M) phase or cell division (cytokinesis) (Fig. 1) (2). The endocycle is a normal developmental cell cycle variation that occurs widely, including in single-celled protists, plants, and humans. A related polyploid cell cycle is known as endomitosis, during which cells enter but do not complete mitosis nor divide (Fig. 1). During most polyploid cell cycles, the genome is duplicated only once per S phase. Rare exceptions to this rule occur in some endocycling cells, where a few gene loci repeatedly rereplicate to support a specific cell function, a process known as developmental gene amplification (3). Much remains unknown about the diversity of cell cycle mechanisms that lead to polyploidy and, in many cases, how an increase in DNA content and cell size supports cell function. Nonetheless, there have been some major advances recently, and I refer the interested reader to two excellent reviews (2, 4).
Fig. 1.
The canonical mitotic cell cycle, endocycle, and endomitosis.
Polyploid Cells Differ in their Completion of Endoreplication
Polyploid cycles have been intensively studied in Drosophila, where cells of many tissues endocycle. Many heterochromatic loci, however, are not duplicated during every endocycle S phase, which results in their progressive underrepresentation relative to fully replicated euchromatic loci (5, 6). Although most of these underreplicated heterochromatic DNA are large blocks of pericentric short repeats, some gene loci with silencing chromatin marks are also underreplicated (7, 8). Evidence indicates that DNA replication is incomplete in all Drosophila polyploid cells examined, but that its extent can differ among loci and tissues (8–10).
In PNAS, Sher et al. use array comparative genomic hybridization to examine two polyploid cell types in mice: endocycling trophoblast giant cells (TGCs) of the placenta, and endomitotic megakaryocytes (MKs), the precursors to blood platelets (1). Their analysis does not reveal developmental amplification of genes that are highly expressed and important for TGC and MK function. Surprisingly, unlike Drosophila, there was also no evidence for incomplete DNA replication. Because the array only contained probes for euchromatic loci, the authors evaluated DNA copy number at three heterochromatic loci by quantitative real-time PCR, which revealed that they are also fully replicated. Although it remains possible that other heterochromatic loci are not fully replicated, these results indicate that polyploidization cycles in TGCs and MKs differ from those in Drosophila, where large segments of the genome are severely underrepresented.
Completing Endoreplication: DNA Replication Genes, Cell Cycle Regulation, and Epigenomic Status
To explore what causes the difference between fly and mouse endoreplication, Sher et al. (1) analyze the expression of protein coding and microRNA genes in TGCs and MKs by microarray and RNA-Seq. A previous study in Drosophila found that many DNA replication genes are expressed at lower levels in endocycling cells than mitotic cycling cells, including genes that encode proteins required at replication origins and forks (11). Strikingly, both TGCs and MKs did not have the same drastic reduction in expression of DNA replication genes, perhaps explaining why DNA replication is more complete in mouse than fly. As Sher et al. (1) discuss, endocycle S phase may be slower in some Drosophila cells than mouse TGCs, and previous comparisons of cells from different Drosophila tissues suggested that the replication fork rate is 10 times slower and S phase up to several-fold longer in endocycling than mitotic cycling cells (12–14). Recent results indicate that underreplicated regions have an even slower replication fork rate and a paucity of active replication origins (15).
These data suggest, therefore, that the proteins that are required at replication origins and forks may be limiting in Drosophila for endoreplication of heterochromatin. Previous data, however, suggested that it is the altered oscillations of Cyclin E/cyclin-dependent kinase 2 (CDK2) activity during endocycles that causes underreplication (9). Heterochromatic loci normally replicate during late S phase of a mitotic division cycle, but during endocycles, Cyclin E/CDK2 activity drops and endocycle S phase ends before these heterochromatic loci are duplicated (9). Therefore, a cogent model is that although the Drosophila endocycle S phase is relatively long in duration, DNA replication is slower, and a drop in Cyclin E/CDK2 activity truncates the S phase before the duplication of late-replicating heterochromatin. Thus, underreplication may be determined by the interplay among replication protein levels, oscillations of CDK2 activity, and the epigenomic status of a locus in a specific cell type. An important test will be to identify which replication proteins are truly limiting for late replication in Drosophila, and determine whether fork rate and origin density differ between fly and mouse polyploid cells.
Mouse and Drosophila Polyploid Cycles: Different but Similar
Although Sher et al. (1) focus on the differences between Drosophila and mouse polyploid cycles, their data also uncover many similarities. One of the most notable similarities is the transcriptional repression of multiple genes required for mitosis and cytokinesis (1, 11, 16, 17). Two recent reports indicated that mouse TGCs and polyploid liver cells both have lower expression of genes required for mitosis and cytokinesis, many of which are also repressed during Drosophila endocycles (11, 18, 19). Sher et al. find that some of the mitotic genes are not as repressed in the endomitotic MKs, suggesting that this may distinguish endocycles from endomitosis (1).
The lower expression of mitotic genes is part of a larger similarity between mouse and Drosophila polyploid cycles: the dampened expression of genes regulated by the E2F family of transcription factors. In Drosophila, this dampened E2F transcriptome includes the DNA replication genes as well as the mitotic and cytokinesis genes (11, 18–21). Unlike TGCs, mouse liver cells also have lower levels of E2F-regulated DNA replication genes that are orthologous to those repressed during Drosophila endocycles (11, 19). This finding raises the question whether the genome is fully duplicated during liver endoreplication. Transcriptome analysis has revealed other similarities between mouse and Drosophila polyploid cycles, including altered expression of genes for metabolism and apoptosis (1, 11, 18, 19). Indeed, mouse TGCs and Drosophila endocycling cells both repress the apoptotic response to DNA damage (22–24). Thus, there are fundamental similarities and differences among polyploid cycles in different cell types of mouse, fly, and other organisms.
Cell Polyploidization: Variations on a Cell Cycle Variation
An open question is whether the difference between insects and mammals that Sher et al. (1) describe will apply to other tissues and organisms. It is possible that other polyploid cell types in mammals underreplicate to some degree, whereas some recently described polyploid cell types of Drosophila may have more complete replication (25, 26). Indeed, genome duplication is complete during the first five endocycles of Drosophila ovarian nurse cells, and some heterochromatic DNA may be fully replicated in salivary glands of other insects (10, 27). Similar to mouse, endocycling leaf cells of Arabidopsis thaliana also fully duplicate their heterochromatic DNA (28). Thus, there is a diversity of endoreplication programs among tissues as well as organisms. The report from Sher et al. (1) is an important advance for understanding this diversity. It remains unclear, however, whether underreplication fulfills a specific purpose or is simply a result of altered regulation of the polyploid cell cycle. Nonetheless, given that some cancer cells inappropriately engage an endocycle program, further investigation into underreplication during development may reveal new mechanisms that contribute to genome instability in the cancer cell (29). The genomic data from Sher et al. (1) is a rich treasure trove for future research into these polyploid cell cycle variations, and demonstrates that when it comes to making big cells, one size certainly does not fit all.
Footnotes
The author declares no conflict of interest.
See companion article on page 9368 of issue 23 in volume 110.
References
- 1.Sher N, et al. Fundamental differences in endoreplication in mammals and Drosophila revealed by analysis of endocycling and endomitotic cells. Proc Natl Acad Sci USA. 2013;110(23):9368–9373. doi: 10.1073/pnas.1304889110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox DT, Duronio RJ. Endoreplication and polyploidy: Insights into development and disease. Development. 2013;140(1):3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvi BR. Developmental DNA amplification. In: DePamphilis ML, editor. DNA Replication and Human Disease. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2006. pp. 233–255. [Google Scholar]
- 4.Nordman J, Orr-Weaver TL. Regulation of DNA replication during development. Development. 2012;139(3):455–464. doi: 10.1242/dev.061838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gall JG, Cohen EH, Polan ML. Reptitive DNA sequences in Drosophila. Chromosoma. 1971;33(3):319–344. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- 6.Smith AV, Orr-Weaver TL. The regulation of the cell cycle during Drosophila embryogenesis: The transition to polyteny. Development. 1991;112(4):997–1008. doi: 10.1242/dev.112.4.997. [DOI] [PubMed] [Google Scholar]
- 7.Belyaeva ES, et al. Late replication domains in polytene and non-polytene cells of Drosophila melanogaster. PLoS ONE. 2012;7(1):e30035. doi: 10.1371/journal.pone.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nordman J, Li S, Eng T, Macalpine D, Orr-Weaver TL. Developmental control of the DNA replication and transcription programs. Genome Res. 2011;21(2):175–181. doi: 10.1101/gr.114611.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lilly MA, Spradling AC. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 1996;10(19):2514–2526. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- 10.Dej KJ, Spradling AC. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development. 1999;126(2):293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- 11.Maqbool SB, et al. Dampened activity of E2F1-DP and Myb-MuvB transcription factors in Drosophila endocycling cells. J Cell Sci. 2010;123(Pt 23):4095–4106. doi: 10.1242/jcs.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skora AD, Spradling AC. Epigenetic stability increases extensively during Drosophila follicle stem cell differentiation. Proc Natl Acad Sci USA. 2010;107(16):7389–7394. doi: 10.1073/pnas.1003180107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierce SB, et al. dMyc is required for larval growth and endoreplication in Drosophila. Development. 2004;131(10):2317–2327. doi: 10.1242/dev.01108. [DOI] [PubMed] [Google Scholar]
- 14.Spradling A, Leys E. Slow replication fork movement during Drosophila chorion gene amplification. In: Kelly T, Stillman B, editors. Cancer Cells. Cold Spring Harbor: Cold Spring Harbor Lab Press; 1988. pp. 305–309. [Google Scholar]
- 15.Sher N, et al. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 2012;22(1):64–75. doi: 10.1101/gr.126003.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narbonne-Reveau K, et al. APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development. 2008;135(8):1451–1461. doi: 10.1242/dev.016295. [DOI] [PubMed] [Google Scholar]
- 17.Zielke N, Querings S, Rottig C, Lehner C, Sprenger F. The anaphase-promoting complex/cyclosome (APC/C) is required for rereplication control in endoreplication cycles. Genes Dev. 2008;22(12):1690–1703. doi: 10.1101/gad.469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandit SK, et al. E2F8 is essential for polyploidization in mammalian cells. Nat Cell Biol. 2012;14(11):1181–1191. doi: 10.1038/ncb2585. [DOI] [PubMed] [Google Scholar]
- 19.Chen HZ, et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat Cell Biol. 2012;14(11):1192–1202. doi: 10.1038/ncb2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng L, Zhu C, Xu J, Du W. Critical role of active repression by E2F and Rb proteins in endoreplication during Drosophila development. EMBO J. 2003;22(15):3865–3875. doi: 10.1093/emboj/cdg373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zielke N, et al. Control of Drosophila endocycles by E2F and CRL4(CDT2) Nature. 2011;480(7375):123–127. doi: 10.1038/nature10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 2008;22(22):3158–3171. doi: 10.1101/gad.1710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ullah Z, Kohn MJ, Yagi R, Vassilev LT, DePamphilis ML. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev. 2008;22(21):3024–3036. doi: 10.1101/gad.1718108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soloveva V, Linzer DI. Differentiation of placental trophoblast giant cells requires downregulation of p53 and Rb. Placenta. 2004;25(1):29–36. doi: 10.1016/S0143-4004(03)00215-7. [DOI] [PubMed] [Google Scholar]
- 25.Fox DT, Gall JG, Spradling AC. Error-prone polyploid mitosis during normal Drosophila development. Genes Dev. 2010;24(20):2294–2302. doi: 10.1101/gad.1952710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unhavaithaya Y, Orr-Weaver TL. Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev. 2012;26(1):31–36. doi: 10.1101/gad.177436.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinemann M. Co-replication of satellite DNA of Chironomus melanotus with mainband DNA during polytenization. Chromosoma. 1978;66(2):127–139. doi: 10.1007/BF00295135. [DOI] [PubMed] [Google Scholar]
- 28.Jacob Y, et al. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature. 2010;466(7309):987–991. doi: 10.1038/nature09290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]