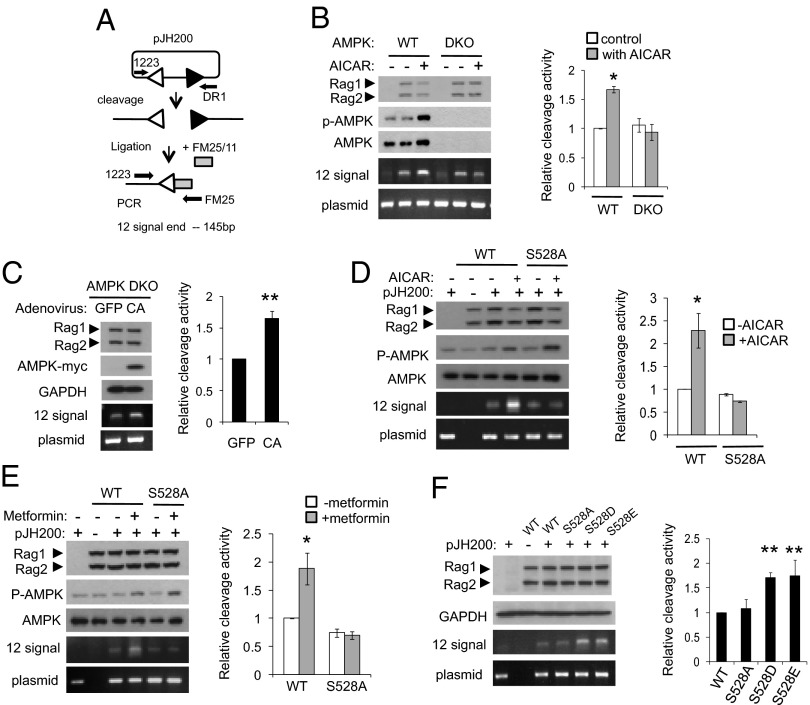
Fig. 4.
Increased cleavage of an extrachromosomal substrate by AMPK activation in vivo. (A) Diagram of the ligation-mediated PCR (LM-PCR) cleavage assay using the extrachromosomal V(D)J substrate pJH200. The linker (FM25/11) is shown as a shaded box. PCR primers 1223 and FM25 were used to amplify the cleaved fragment. To control for recovery of the recombination substrate, primers 1223 and DR1 were used to amplify a portion of the backbone of the plasmid (39, 40). (B) MBP-RAG1, MBP-RAG2, and pJH200 were transiently transfected in WT MEF cells or in AMPK DKO MEF cells. Cells were treated with 2 mM AICAR for 3 h. Cleavage was detected by LM-PCR. A representative experiment is shown (Left). Relative activities were quantified from at least three independent experiments (Right). Results are mean ± SE. *P < 0.05 between control and AICAR-treated cells. (C) Signal ends were detected in AMPK DKO cells expressing RAG1, RAG2, and an adenovirally expressed constitutively active form of AMPK (CA) or GFP. The level of the signal end was normalized to the level of the plasmid backbone and quantified from three independent experiments. Results are mean ± SE. **P < 0.001 between GFP and CA. (D and E) WT or S528A RAG1, RAG2, and pJH200 were cotransfected in 293T cells and treated with either 2 mM AICAR for 3 h (D) or 1 mM metformin for 3 h (E). Signal ends were detected by LM-PCR (Left). Relative amount of signal end was normalized with that of the unrecombined plasmid and quantified from three independent experiments (Right). Results are mean ± SE. *P < 0.05. (F) 293T cells were cotransfected with MBP-WT, S528A, S528D, or S528E RAG1, MBP-RAG2, and pJH200. Signal ends were detected by LM-PCR (Left). Relative activities were quantified from at least three independent experiments (Right). Results are mean ± SE. **P < 0.001 between WT and S528 mutants.