Figure 5. Bridging in biological capsids. Number of NA segments that directly interact with positively charged ARM segments (interaction energy , blue squares) and bridging segments (interaction energy , purple circles).
The numbers are calculated at the optimal length for each capsid shown in Figure 4 using the base-paired model. For visual reference, the dashed line indicates a 1:1 correspondence between capsid charge and number of nucleotides.
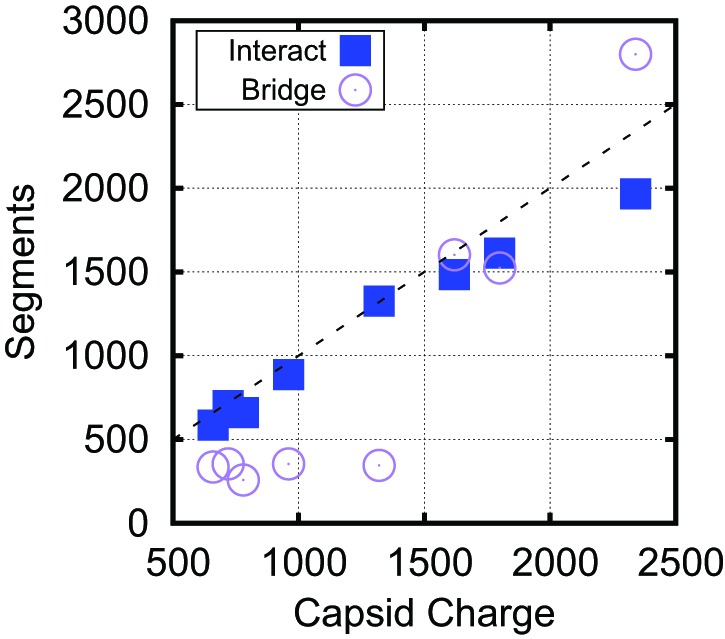
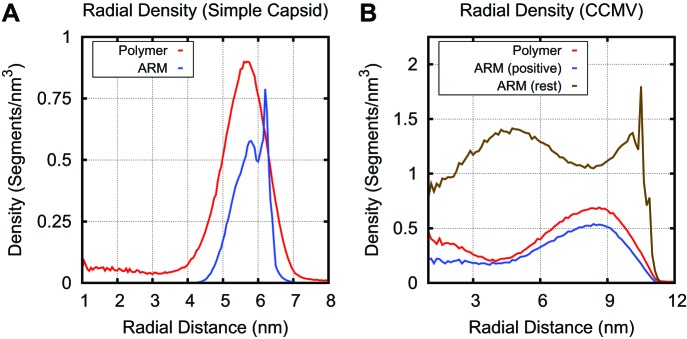
Figure 5—figure supplement 1. Radial density for linear polymer and ARM segments in the simple capsid (A) and CCMV (B).
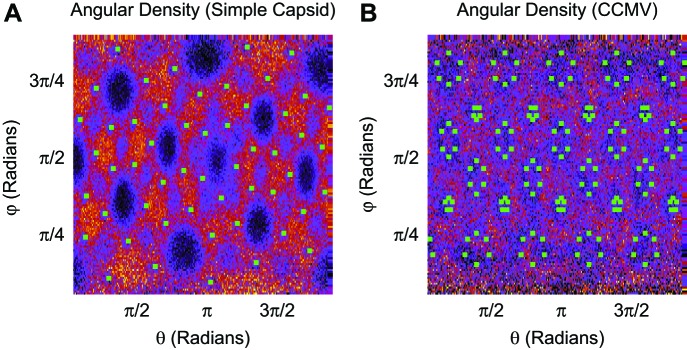
Figure 5—figure supplement 2. Angular density of linear polymer segments (heat map) in the basic capsid model (A) and CCMV (B).
Figure 5—figure supplement 3. Capsid radius and polymer bridging.

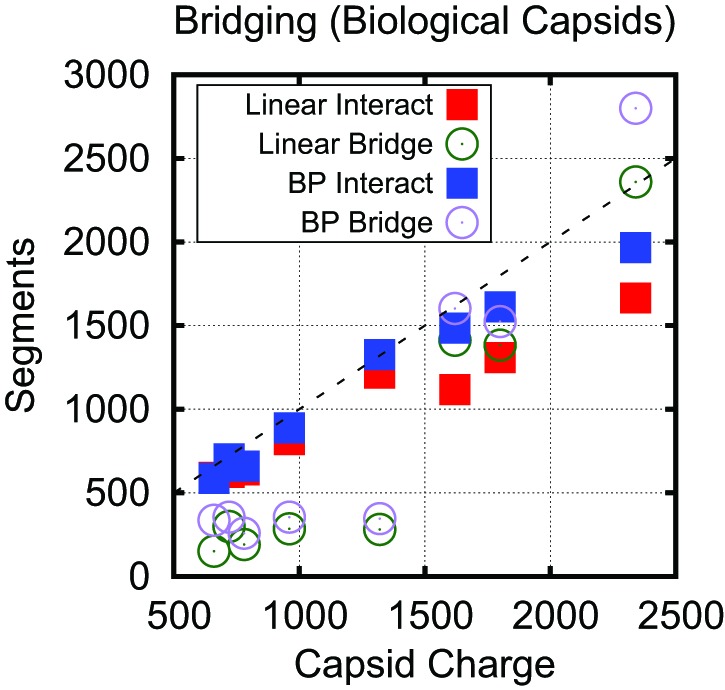
Figure 5—figure supplement 4. Number of NA segments that directly interact with positively charged ARM segments and bridging segments, for both the linear and base-paired model.