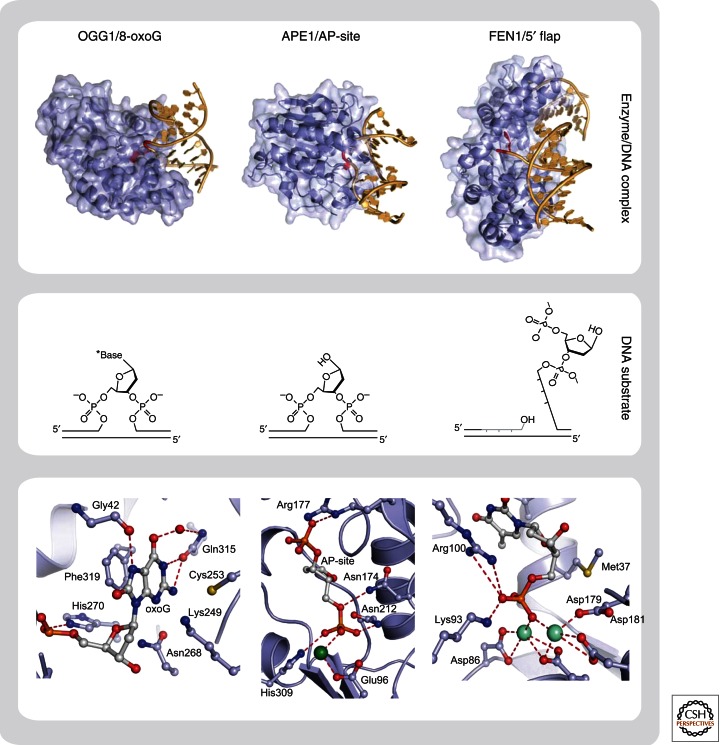
Figure 3.
Structural basis for interaction of BER enzymes with their DNA substrates. Overall structure (upper panel) and close-up of active site (lower panel) of three different repair proteins in complex with their DNA substrates. Recognition and extrahelical flipping of damaged base (8-oxoG) by DNA glycosylase (OGG1) (Radom et al. 2007), PDB code 2NOZ; binding of AP-site by AP-endonuclease (APE1) (Mol et al. 2000), PDB code 1DE8; and recognition of 5′ flap by structure specific endonuclease (FEN1) (Tsutakawa et al. 2011), PDB code 3Q8K. The chemical structures of DNA substrates are shown in the middle panel.