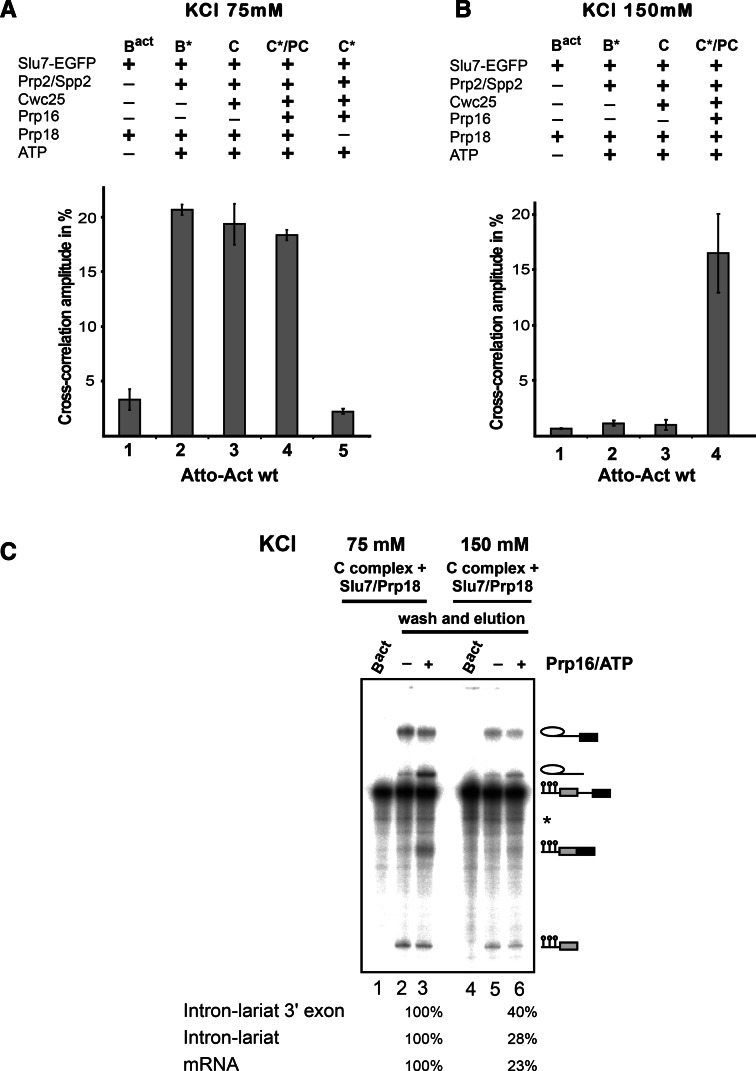
FIGURE 4.
A low-affinity binding site is created for Slu7-EGFP during catalytic activation of the spliceosome by Prp2 as analyzed by dcFCCS. (A) Affinity-purified Bact complexes assembled on Atto-Actwt were complemented on the matrix with recombinant Slu7-EGFP plus Prp18 (column 1, Bact) and additionally with Prp2 and Spp2 (column 2, B*), Cwc25 (column 3, C), Prp16 (column 4, C*/PC), or without Prp18 (column 5, C*). C*/PC indicates that upon Slu7/Prp18 binding to the C* complex, the latter is transformed into the post-catalytic complex (PC). After incubation, complexes were washed with GK75 buffer or GK150 buffer (B) and then eluted from the matrix with maltose. dcFCCS measurements were then performed at complex concentrations of 1.0 nM. Cross-correlation amplitudes derived from two independent experiments are shown for each complex. Error bars indicate the standard deviation from two independent measurements. (C) Reconstituted C complexes were incubated on the matrix with Slu7/Prp18 at 75 or 150 mM KCl, respectively. Complexes were washed with buffers containing 75 or 150 mM KCl, eluted from the matrix in GK75 buffer and mock-treated (lanes 2,5) or incubated in solution with Prp16 plus ATP (lanes 3,6). After incubation, RNAs were extracted, separated by denaturing PAGE, and visualized by autoradiography. Symbols for RNA species (as in Fig. 1) are shown on the right. The intensities of the intermediate and mRNA product signals were quantified using the ImageQuant software (Molecular Dynamics). The signals in lane 3 were used as 100%.