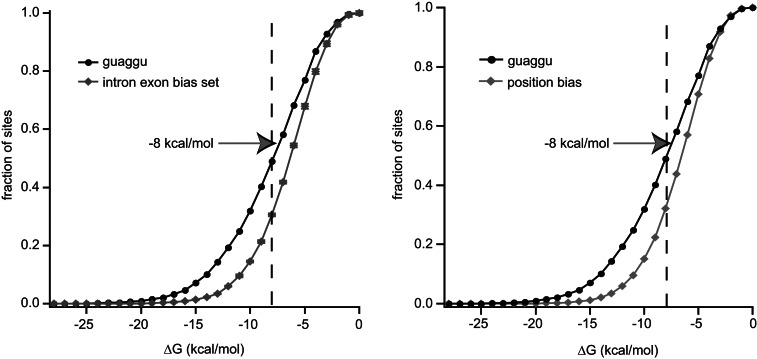
FIGURE 5.
GUAGGU 5′ splice sites are located in predicted secondary structure-rich sequences. (Left) The 39-nucleotide sequences surrounding the 5′ splice site of exons with a GUAGGU 5′ splice site or three sets of permuted artificial exon sequences were folded using the Vienna RNA package. The fraction of sequences folding at a given energy level was plotted versus the predicted free energy of folding. Error bars on the permuted set indicate the standard deviation of the three artificial sets. See the Materials and Methods for details regarding the construction of this set. A dashed line indicates the position of −8 kcal/mol. (Right) The predicted free energy of folding of GUAGGU exons was plotted against a background set of 100,000 random 39 mers, in which the nucleotide bias of each individual position relative to the 5′ splice site was preserved.