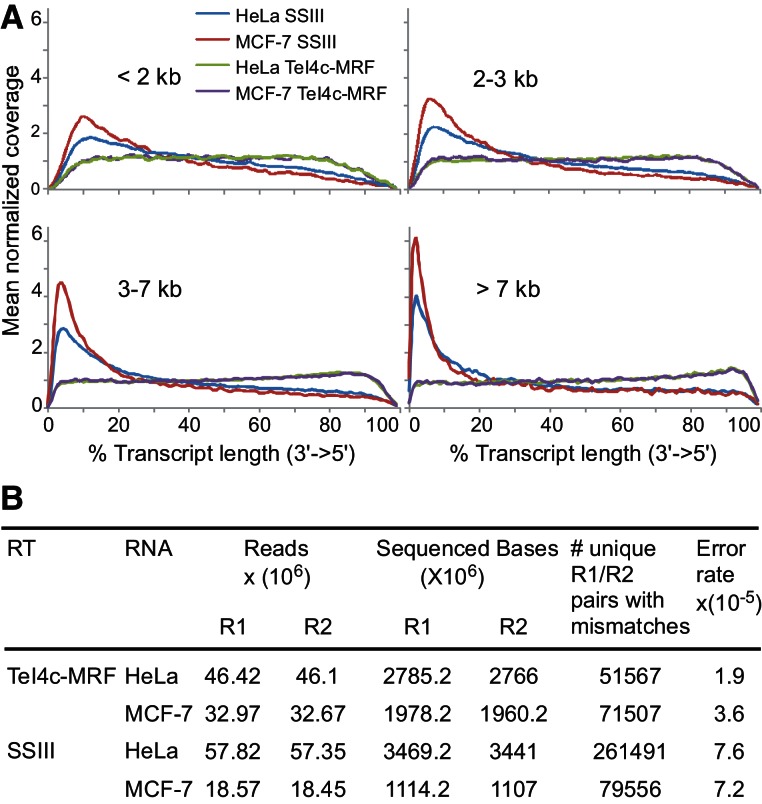
FIGURE 5.
RNA-seq with a group II intron and retroviral RT. HeLa or MCF-7 RNAs were annealed with an oligo(dT)42 primer and incubated with TeI4c-MRF RT (1.24 μM) at 60°C or SuperScript III (10 units/μL) for 2 h at 50°C. The cDNAs were converted into RNA-seq libraries and paired-end sequenced on an Illumina HiSeq. (A) Distribution of reads per unit length for transcripts of different size classes. Reads were aligned using Eland-32 to a set of ∼7203 transcripts curated by selecting the longest isoform of each annotated gene from RefSeq (downloaded 11/2010), removing sequences containing ambiguous bases, and requiring that >95% of bases could be uniquely mapped to RefSeq and have mean base coverage >3X in standard brain and/or UHR mRNA data sets. (B) Error frequencies. Raw data were base-called using the Illumina Off-Line Basecaller (OLB version 1.9), and the resulting reads were aligned to human NCBI reference build 36 and splice junctions from UCSC refFlat (downloaded 02/2010) using Eland RNA (Casava 1.7) with default parameters. Potential RT errors were detected by looking for single-base mismatches relative to the reference sequence in overlapping sequence in both reads R1 and R2 of a paired-end cluster. Both R1 and R2 were required to have a base quality >25 and belong to a perfectly overlapping section of length ≥20 nt. Base mismatches common to both the TeI4c-MRF and SuperScript III libraries, which include sequence polymorphisms compared with the reference RNAs, were filtered out.