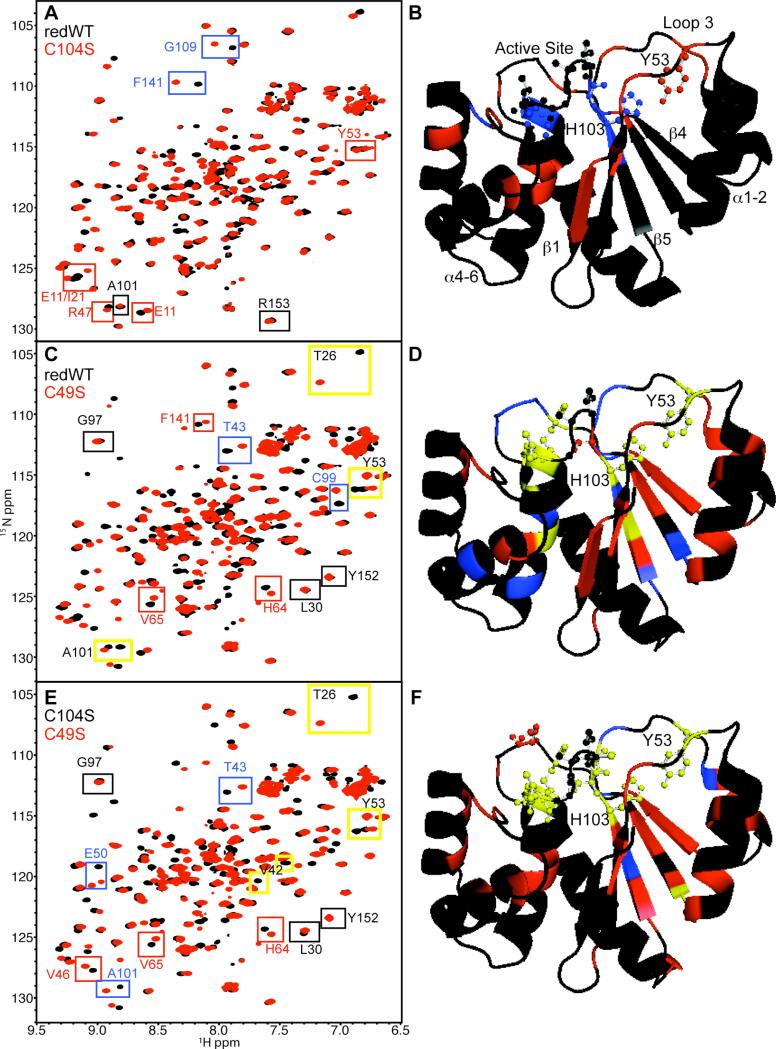
Figure 5. Chemical Shift Mapping of PRL-1.
A. 1H-15N HSQC spectra of reduced of wild type (black) and C104S (red) are shown. Chemical shift changes between the two spectra were analyzed as described in the methods section. A representative example from each category described below is boxed in the spectra. Black boxes highlight residues with Δf < 33.8. Red boxes emphasize residues with 33.8 < Δf < 100 Hz. Blue boxes specify residues with 300 Hz < Δf > 100 Hz. B. Chemical shift changes (Δf) between reduced wild type and C104S were mapped to the available C104S (PDB 1XM2)36 crystal structure and color-coded as in A. The active site, Y53 and H103 residues are depicted as ball and sticks. Relevant regions of the protein are also labeled. C. 1H-15N HSQC spectra of reduced wild type (black) and C49S (red) with colored boxes highlighting chemical shift change from the corresponding categories. Additionally, residues with Δf > 300 Hz are specified by yellow boxes with black lettering. D. As in B except changes between the reduced wild type and C49S are mapped. The same chemical shift change categories used in panel C were used in panel D (no change=black; small change=red; medium change=blue; large change=yellow) E. 1H-15N HSQC spectra of C104S (black) and C49S (red) with colored boxes highlighting chemical shift changes from the corresponding categories. F. As in B and D except chemical shift changes between C104S and C49S were mapped. The same color-code applies. Panels B, E and F were generated using Pymol.