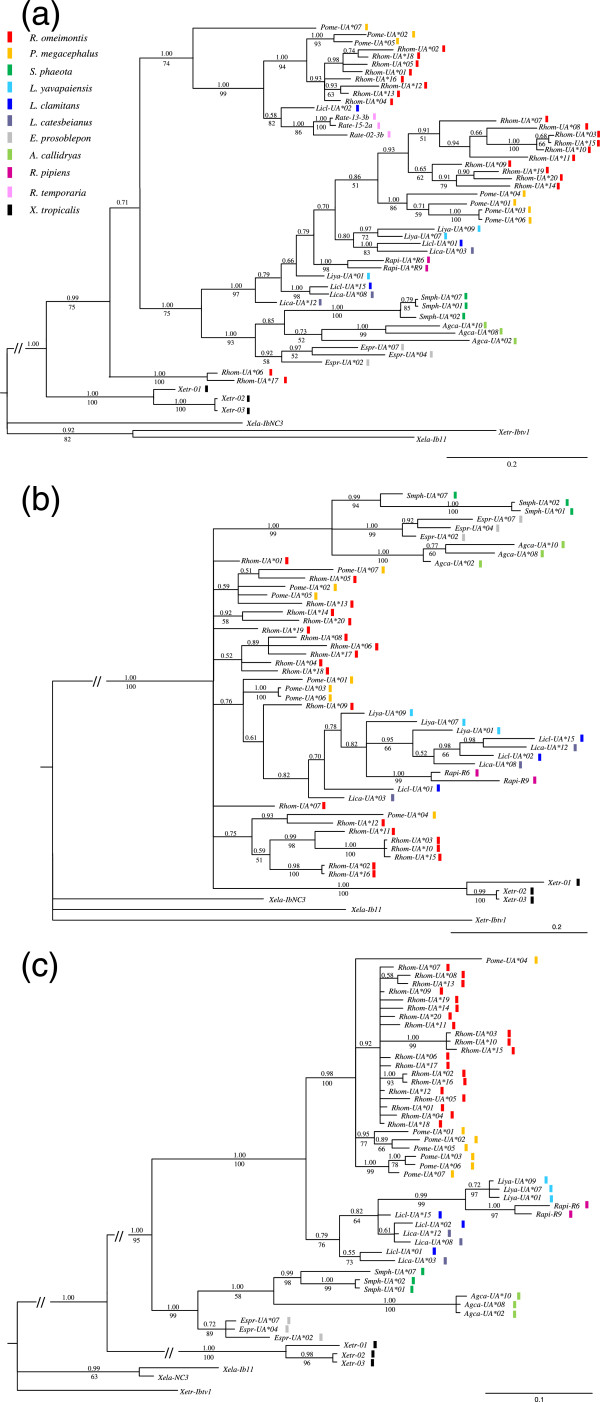
Figure 3.
Phylogenetic relationships of exon 2 (a), exon 3 (b), and exon 4 (c) in anuran MHC class I alleles. All of the trees were rooted from MHC Ib genes of Xenopus. Bootstrap values >50% (from the neighbor-joining analysis) are shown above the respective clades. Bayesian posterior probabilities >0.70 are presented below the clades. All of the branches are proportional to the scale shown at the bottom right of the figure, excluding branches with the sign “//”, which were shortened for graphical clarity of the remaining branches of the tree. Alleles belonging to different species are marked using different colored bars. Involved allelic sequences obtained from NCBI include the following: MHC class Ia alleles from S. phaeota (Smph-UA*07: JQ679386; Smph-UA*02: JQ679381; Smph-UA*01: JQ679380); L. yavapaiensis (Liya-UA*09: JQ679379; Liya-UA*07: JQ679377; Liya-UA*01: JQ679371); L. clamitans (Licl-UA*15: JQ679369; Licl-UA*02: JQ679356; Licl-UA*01: JQ679355); L. catesbeianus (Lica-UA*12: JQ679354; Lica-UA*08: JQ679350; Lica-UA*03: JQ679345); E. prosoblepon (Espr-UA*07: JQ679337; Espr-UA*04: JQ679334; Espr-UA*02: JQ679332); A. callidryas (Agca-UA*10: JQ679321; Agca-UA*08: JQ679319; Agca-UA*02: JQ679313); R. pipiens (Rapi-R6: AF185587; Rapi-R9: AF185588); R. temporaria (Rate-13-3b: FJ385632; Rate-02-3b: FJ385633; Rate-15-2a: FJ385641); X. tropicalis (Xetr-01: BC161748; Xetr-02: NM_001113065; Xetr-03: BC154904); and MHC Ib alleles from X. tropicalis (Xetr-Ibtv1: NM_001247995) and from X. laevis (Xela-Ib11: FJ589643; Xela-IbNC3: L20726).