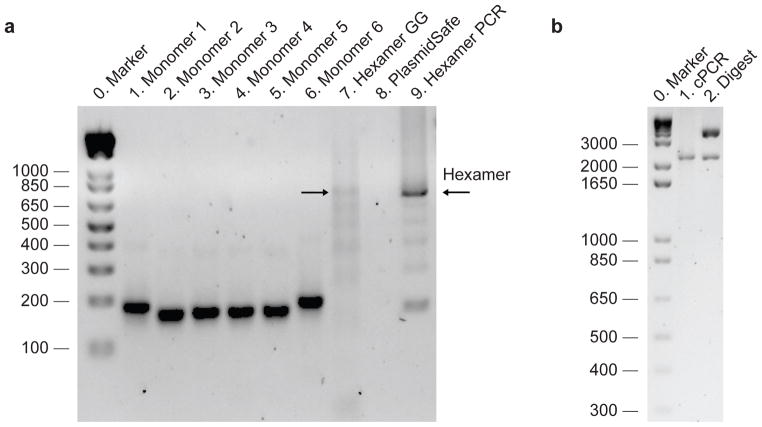
Figure 5. Example gel results from the TALE construction procedure.
(a) Lanes 1—6: products from the monomer PCR reaction (Stage 1 in Figure 3) after purification and gel normalization (Procedure Steps 8–9). The molar concentrations of samples shown on this gel have been normalized so that equal moles of monomers are mixed for downstream steps. Monomers 1 and 6 are slightly longer than monomers 2, 3, 4, and 5 due to the addition of sequences used for circularization. Lane 7: result of the hexamer Golden Gate cut-ligation (Procedure Step 15). A series of bands with size ~700 bp and lower can be seen. Successful hexamer Golden Gate assembly should show a band ~700 bp (as indicated by arrow). Lane 8: hexamer assembly after PlasmidSafe exonuclease treatment (Procedure Step 17). Typically the amount of circular DNA remaining is difficult to visualize by gel. Lane 9: result of hexamer amplification (Procedure Step 20). A ~700 bp band should be clearly visible. The hexamer gel band should be gel-purified to remove shorter DNA fragments. (b) Properly assembled TALE-TFs and TALENs can be verified using bacterial colony PCR (2175 bp band, lane 1) (Procedure Step 35) and restriction digest with AfeI (2118 bp band for correctly assembled 18-mer in either backbone; other bands for TALE-TF are 165 bp, 3435 bp, 3544 bp; other bands for TALEN are 165 bp, 2803 bp, 3236 bp; digest shown is for TALE-TF backbone vector, lane 2) (Procedure Step 35).