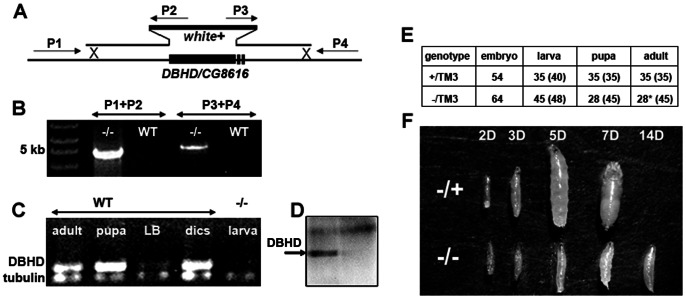
Figure 1. Generating a DBHD null allele.
(A) The DBHD genomic locus and the targeting strategy. P1–P4 represent the PCR primers. (B) PCR analysis of the genomic DNA. In DBHD−/− larvae (−/−), a 4.8 kb fragment and a 5 kb fragment could be amplified by the corresponding primer pairs. (C) rtPCR analysis of the DBHD transcripts in various tissues. LB: larval brain; disc: mixtures of larval imaginal discs. α-tubulin (at 84B) was used as the positive control. (D) A DBHD antibody recognizes a band at about 55 kDa (arrow) of the whole larval extracts, which is absent in the DBHD−/− larvae. w1118 was used as the wild-type control (WT). (E) Statistical analysis of the developmental profiles of two strains. The +/TM3 flies contains a healthy 3rd chromosome and a GFP-marked third chromosome balancer (TM3, Kr::GFP). The −/TM3 flies contains the same balancer and the DBHD− allele. Animals survived to different stages were counted. Numbers in the parenthesis are the theoretical values according to the Mendel rules. *: all the survived adults are heterozygotes. (F) Comparison of DBHD−/− (−/−) and the sibling heterozygotes (−/+) at different days after egg laying. Embryos collected within three hours and cultured in the same food vials were picked for images at each time point. All heterozygotes have eclosed by 14 days after egg laying and thus were not pictured.