Abstract
Background
Age is the strongest breast cancer risk factor, with overall breast cancer risk increasing steadily beginning at approximately 30 years of age. However, while breast cancer risk is lower among younger women, young women's breast cancer may be more aggressive. Though several genomic and epidemiologic studies have shown higher prevalence of aggressive, estrogen-receptor negative breast cancer in younger women, the age-related gene expression that predisposes to these tumors is poorly understood. Characterizing age-related patterns of gene expression in normal breast tissues may provide insights on etiology of distinct breast cancer subtypes that arise from these tissues.
Methods
To identify age-related changes in normal breast tissue, 96 tissue specimens from reduction mammoplasty patients aged 14 to 70 were assayed by gene expression microarray.
Results
Significant associations between gene expression levels and age were identified for 802 probes (481 increased, 321 decreased with increasing age). Enriched functions included ‘aging of cells’, ‘shape change’, and ‘chemotaxis’, and enriched pathways included Wnt/beta-catenin signaling, Ephrin Receptor Signaling, and JAK/Stat Signaling. Applying the age-associated genes to publicly available tumor datasets, the age-associated pathways defined two groups of tumors with distinct survival.
Conclusion
The hazard rates of young-like tumors mirrored that of high grade tumors in the Surveillance, Epidemiology and End Results Program, providing a biological link between normal aging and age-related tumor aggressiveness.
Impact
These data show that studies of normal tissue gene expression can yield important insights about the pathways and biological pressures that are relevant during tumor etiology and progression.
Keywords: aging, breast, cancer, gene expression, reduction mammoplasty
Introduction
Age is the strongest demographic risk factor for human cancer overall (1, 2), with breast cancer rates steadily increasing with age. However, while tumors are less common in young women, younger women are more likely to have aggressive tumors. Young women's breast cancer is more often estrogen receptor (ER) negative, while older women's cancers are more often ER-positive (3, 4). Two previous gene expression studies have compared molecular features of breast cancer by age, focusing on the tumor gene expression (5, 6). These studies have shown that young women's tumors have distinct gene expression, ultimately reflecting different prevalence of breast cancer subtypes. In some analyses, age-associated gene expression changes were no longer evident after adjusting for subtype and grade (7). Thus, persistent gaps in our understanding of age-associated changes in tumor aggressiveness remain.
Research is needed to distinguish characteristics of the malignancy from those of the host (5, 8) and to distinguish background effects of aging from those that contribute to carcinogenesis (9). Though the host-tumor interface has important consequences for a nascent tumor (10-12), changes to this interface with aging are poorly understood (7). Beyond the histology of postmenopausal involution, aging of normal breast tissue has had limited study (13), and gene expression studies of normal tissue may provide important insights.
Normal tissue studies can help to identify barriers that must be overcome by tumor cells (14). Barriers to carcinogenesis derive from normal tissue homeostasis in the microenvironment prior to, or early in, disease (15). Because tumors evolve with selective pressure from surrounding stroma, studies of normal breast tissue, typically more than 90% stroma by volume, may provide insights regarding selective pressures faced by tumors. We hypothesized that older vs. younger tissues represent distinct evolutionary environments, resulting in distinct cancer subtypes by age. Under this hypothesis, gene expression in the normal tissue of young women would be reflected in aggressive tumors common in young women. To evaluate this hypothesis, we identified age-associated gene expression in 96 normal tissues from premenopausal reduction mammoplasty (RM) patients and tested this signature in independent normal breast microarray data. We then evaluated the age-associated signature using publicly available tumor tissue gene expression data, asking whether age-associated gene expression from normal defines distinct tumor groups. The results link gene expression in young women's breast tissue with aggressive tumor phenotypes.
Methods
Patient characteristics
This study included women age 14 – 70 who underwent RM surgery at Baystate Medical Center in Springfield, Massachusetts between 2007 and 2009. Patient characteristics are presented in Table 1. Institutional Review Boards (IRBs) at Baystate and University of Massachusetts Amherst approved the study. Women consented to provide tissues not needed for diagnostic purposes to complete a telephone interview following surgery. Tissues were snap frozen and stored at -80° C. An independent data set of isolated glands from RMs was from UCSF Cancer Center and the Cooperative Human Tissue Network, with patients consented under an Institutional Review Board at those institutions.
Table 1. Demographic characteristics of women*.
| Cases | ||
|---|---|---|
| N | % | |
|
|
||
| Menopausal Status† | ||
| Premenopausal‡ | 62 | 81.6 |
| Perimenopausal§ | 14 | 18.4 |
| Missing | 0 | |
| Race & Hispanic Ethnicity | ||
| White, non-Hispanic | 48 | 63.2 |
| Black, non-Hispanic | 6 | 7.9 |
| Hispanic | 19 | 25.0 |
| Other | 3 | 3.9 |
| Missing | 0 | |
| Age# | ||
| 20-29 | 20 | 26.3 |
| 30-39 | 33 | 43.4 |
| 40-49 | 17 | 22.4 |
| >=50 | 6 | 7.9 |
| Missing | 0 | |
| * Only included individuals >= 20 who are not menopausal | ||
| † 23 women who were postmenopausal were analyzed in pre/peri vs. postmenopausal analyses, but excluded from age analyses and from Table 1. | ||
| ‡ Premenopausal if reports regular periods, or very young age on exogenous hormones, or very young age with hysterectomy, both ovaries preserved | ||
| § Perimenopausal if last menstrual period < 1 year before interview | ||
| # excluded are those < 20 years of age. | ||
| Table 1a: Average Body Mass Index (BMI) by age category | ||
|---|---|---|
| N | BMI | |
|
|
||
| Age | ||
| 20-29 | 15 | 30.4 |
| 30-39 | 16 | 29.4 |
| 40-49 | 13 | 30.1 |
| >= 50 | 2 | 30.5 |
| Missing | 30 | |
RNA isolation and microarrays
Isolation of RNA from RM tissue was performed as described in Sun et al. (16). Agilent 4X44K V1 and custom 244k arrays were performed according to manufacturer protocols for linear amplification and two-color hybridization. Spots that had an intensity of greater than 10 in at least 80% of samples were selected for subsequent analyses. Data were lowess normalized and missing data were imputed using k-nearest neighbors with k=10. Duplicate arrays (N=15) were average. A total of 114 microarrays representing 99 patients were included (GSE33526 for n=72, GEO Submission in progress for n=42). All statistical analyses were performed in R, Bioconductor. For age-associated and menopause-associated gene expression analyses (n=76 and n=99, respectively), probes with no Entrez Gene ID or with less than median variability were eliminated. Linear regression with LIMMA (17) was used to identify significant probes associated with chronological age (in decades) or menopausal status (pre/peri-menopausal vs. postmenopausal). Unadjusted p-values were used with qvalue (Bioconductor) to identify q-values < 0.10 (defined as statistical significance). Hierarchical clustering was used to visualize gene expression, with samples ordered by chronological age and genes clustered by Pearson correlation. Gene ontology analyses were performed using Ingenuity Pathway Analysis (IPA, Redwood City, CA).
We tested a second dataset of isolated glands from RM patients (N=30). Tissue samples were minced and enzymatically dissociated using 0.1% w/v collagenase I in Dulbecco's Modified Eagle Medium at 37 °C for 12 to 18 h. Organoids remaining after digestion were collected by centrifugation at 100 g for two minutes and stored frozen. RNA was extracted using Qiagen RNeasy and Affymetrix GeneChip Human Gene 1.0 ST microarrays were performed at University of Wisconsin, Madison. Microarray data was processed using Robust Multiarray Average (Bioconductor).
To confirm expression changes for CDKN2A and TP53 as identified in microarray data, 1 ug RNA per sample for 26 samples were reverse transcribed using QuantiTect Reverse Transcription kit (Qiagen). Resultant cDNAs (10 ng) were analyzed in triplicate (using miScript SYBR Green PCR Kit and miScript-derived primers for p16 (CDK2NA), p53 or GAPDH) by real-time, quantitative PCR on an Applied Biosystems 7900HT thermocycler. An outlier was detected in regression diagnostics and removed to yield a final QPCR datasets (n=25).
Public Microarray Datasets and Test Set of Isolated Glands
Based on the previous evidence that increasing age is associated with qualitative shifts in tumor subtype (5, 18), we hypothesized that gene expression of normal aging would be manifest in tumors, with more aggressive tumors similar to younger, normal tissue. To test this, we projected the age-associated gene set onto publicly available microarray datasets: the NKI295 (19), Naderi et al. (20), and UNC337 (21). To classify tumors as young-like or older-like, we applied methods described in Creighton et al. (22) to obtain a correlation coefficient describing the relation between each sample and the 802-probe age associated signature. Probes were collapsed by statistical mean to a list of 719 unique entrez genes. The vector summarizing the age signature on these genes was constructed as median expression in RM patients younger than 30 minus that in patients older than 39. Genes with differences less than zero (lower expression in young) were set to -1 and genes with differences larger than zero (higher expression in young) were set to 1. This vector was compared with the sample gene expression data to calculate Pearson correlation coefficients. If the correlation was positive, the patient was classified as young-like; if correlation was negative, the patient was classified as older-like. Since all patients in the RM data set and the NKI dataset were 55 or younger, all three tumor datasets were restricted to this age range for combined analysis. These data were median-centered by gene and filtered to include only those genes with top 50% variability prior to correlation analysis. The 719 genes mapped to 380 variable genes in the NKI dataset, 317 variable genes in Naderi, and 268 variable genes in UNC337. Following classification, all samples were aggregated to a single dataset (n=459).
To test whether young-like tumors mirror aggressive tumors, we used data for women 55 or younger in the Surveillance, Epidemiology, and End Results (SEER) Program (23). Hazard curves for public microarray data and SEER data were generated using the muhaz library in R. Young-like and older-like tumor (microarray data) hazard rates was compared with ‘aggressive’ (grade 3) tumors and ‘less aggressive’ tumors (grade 1 or 2) in SEER. Previous manuscripts (3) have demonstrated that several clinical characteristics (ER status, PR status, race, grade, tumor size) duplicate the same general patterns for age at incidence and hazard rate, so grade was representative.
Creighton correlation-based classifications were also used for analyzing age-associated gene expression in a test set of isolated glands. Entrez IDs from the age signature were mapped to the University of Wisconsin data, with 138 probes identified in common. The association between the Creighton correlation (coded as ‘positive’ if >= 0 or ‘negative’ if < 0) and the true age of tissue (< 30, 30-39, >39) was estimated using the nonzero correlation statistic (1 degree-of-freedom Cochran-Mantel-Haenszel statistic, PROC FREQ in SAS 9.2).
Comparison of Age-Associated Signature with Previously Published Signatures
Previously, a signature of aging based on tumor gene expression data (5) and a meta-analysis of common signatures of aging across many tissues were published (24). These signatures were evaluated in our RM samples to test whether they predicted age in normal human breast tissue and to assess correlation with our aging signature. These signatures were mapped to our filtered RM dataset, resulting in 85 genes for the 145-gene Yau et al. (5) signature and 52 genes for the 73-gene de Magalhaes signature (24). A Creighton correlation was computed (genes with high expression in young coded as 1, high expression in older coded as -1). Positive correlation indicated patients were young-like for a given signature. These classes were then evaluated for association with our RM-based young-like and older-like signature and in association with chronological age. Chi-square tests, or Fisher exact where cell counts were <5, were used to test statistical significance.
Results
Age-associated gene expression in reduction mammoplasty patients
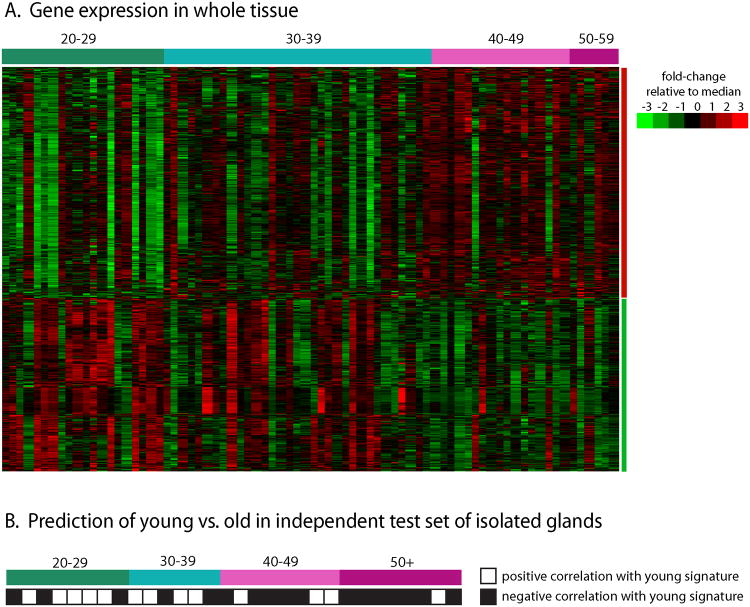
A substantial proportion of genes had expression changes associated with age. Given a False Discovery Rate (FDR) of 5 or 10%, 2 or 4% of genes were age-associated, respectively. A striking 15% of genes were age-associated at FDR of 20%, demonstrating broad changes induced by aging. Figure 1A shows a one-dimensional (gene only) cluster for 802 age-associated genes (with FDR<10%) across 76 samples (representing 62 patients), ordered by chronological age. A qualitative shift is evident in gene expression in the late thirties, with substantial inter-individual variation.
Figure 1. Age-associated gene expression in whole tissue (n=76) and in isolated glands (n=30).
(A) 802 probes were collapsed to unique genes by averaging and a hierarchical cluster analysis (genes-only) shows two distinct gene expression groups. Some heterogeneity is observed within groups, but an overall trend is evident. (B) The gene expression signature was then used to predict age in an independent test set of isolated glands. Glands from younger patients were more likely to show positive correlation with the young signature from whole glands, while older patients were more likely to have negative correlations.
To test this gene set, we used data from isolated glands (enriched for epithelium) of RM patients. Results in this independent test set shows that even in epithelium, the age-related changes observed in whole tissue are preserved. Figure 1B shows a strong correlation between age and the aging signature, with young women positively correlated with the younger women's signature (9 of 14 samples positively correlated) and older women (12 of 16) negatively correlated with young signature. There was a trend toward decreasing correlation with the young-like signature as age increased (odds ratio for young expression was 5.4 [95% CI: 1.1-26.0] comparing younger to older). The association is particularly striking given distinct specimen processing methods and microarray platforms. These data suggest that our signature reflects aging in multiple cell types.
Among the 802 differentially expressed probes, several processes and pathways were over-represented (Table 2). Aging of cells, cell flattening, and shape change were significant processes, while JAK2-associatied hormone-like cytokine signaling, Wnt/β-catenin signaling, and Ephrin Receptor Signaling were significant pathways. The complete list of genes from Figure 1 are in Supplemental Table 1 with the average fold change comparing youngest (< 30, n=20) to oldest patients (>49, n=6).
Table 2. Gene Ontology Categories Enriched Among Age-Associated Genes.
| Ingenuity Category | p-valuea | Molecules |
|---|---|---|
| Functions Annotation | ||
|
|
||
| Aging of cells | 9.4e-5 | NOX4, PDCD4, SOD1, TBX2, TP53 |
| Cell flattening of cell lines | 9.4e-5 | CDKN2A, FIS1, PLD1, SMARCA4, SMARCB1 |
| Shape change | 1.2e-4 | ANGPT1, AP1S2, ARFIP2, ARHGAP15, CASP10, CD28, CDC42EP4, CDKN2A, DLC1, EFNA1, EPHA2, EPHB4, FADD, FIS1, KITLG, MARK2, MYH9, NEDD4L, PACSIN2, PLD1, PLXNB1, RAP1B, RAP2C, ROCK2, RPS6KB1, SEMA3A, SLIT2, SMARCA4, SMARCB1, TEK, TNFSF11 |
| Developmental process of melanocytes | 3.2e-4 | CTNNB1, EFNA1, KITLG, MITF, TP53 |
| Nectortizing enterocolitis | 3.2e-4 | PDE11A, PDE3B, PDE4A, PDE4D, TLR4 |
| Assembly of cellular protrusion | 5.8e-4 | ARFIP2, CCDC88A, RALA, RHOQ, SLC9A3R1, SLIT2 |
| Retraction of cellular protrusion | 6.6e-4 | ARHGAP32, EFNA1, IL6R, ITGA3, MYH9, RHOQ, ROCK2 |
| Canonical Pathways | ||
|
|
||
| Role of JAK2 in Hormone-Like Cytokine Signaling | 1.1e-3 | SHC1, STAT5A, GHR, TYK2, SOCS2, JAK2 |
| Wnt/β-catenin Signaling | 2.9e-3 | TP53, SOX4, CDKN2A, SOX12, MARK2, SOX17, CSNK1E, FZD8, FZD4, SOX8, DVL2, CTNNB1, ACVR1C, WNT5B, SOX5 |
| Ephrin Receptor Signaling | 5.2e-3 | RAP1B, ACTR2, EPHB4, ANGPT1, GNB2L1, LIMK2, JAK2, ITGA3, GNG7, GNG10, EFNA1, ROCK2, SHC1, SDCBP, GRIN2C, EPHA2 |
Benjamini-Hochberg, multiple testing adjusted p-value.
We confirmed the direction of change for CDKN2A (p16) and TP53 by performing Q-RT-PCR on a subset of samples with remaining RNA (Supplemental Figure 1). The p16INK4a tumor suppressor has an established role in aging and its expression increases with increasing age (25, 26). p53 plays a critical role in determining cellular senescence and in vitro lifespan (27) and has declining activity in aging rodents (28). QPCR showed p53 levels decrease with age and p16 levels increase, consistent with the microarray data.
Menopause-associated gene expression in reduction mammoplasty patients
In contrast to a broad gene expression response to age, there were few genes associated with menopausal status. In comparing 76 pre/peri-menopausal to 23 post-menopausal women, only 273 genes were statistically significant with an unadjusted p-value <0.05. No genes were significantly associated with menopausal status after correction for multiple testing (q value > 0.10 for all genes). Despite the weak association, we performed an IPA analysis with the 273 genes that had p-value < 0.05. No Functional Annotation or Canonical Pathway categories were differentially expressed with Benjamini-Hochberg p<0.05. These results show that within the age range 20-70, menopausal status did not strongly influence breast tissue gene expression.
Age-associated gene expression in the breast cancer patients
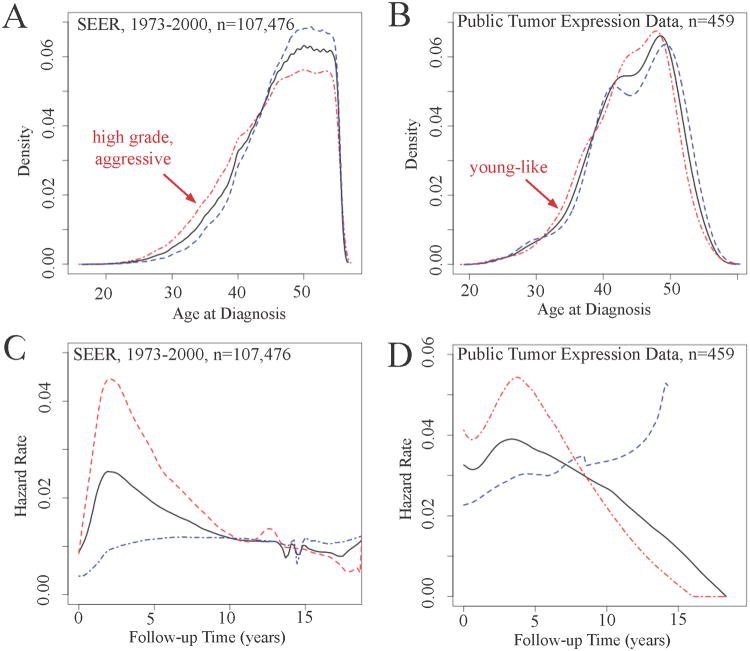
According to evolutionary theories of cancer (11), tumors use the transcriptional programs and pathways that are active in normal tissue to advance growth and survival. Thus we expected that age-associated genes in normal tissue would also be dysregulated in tumors. By applying the age-associated gene set from Figure 1 to three public microarray datasets, we identified two groups of patients. ‘Aggressive’ high grade patients in SEER (Figure 2A) and patients with young-like gene expression (Figure 2B) both showed a left shift in the age-at-incidence distribution, documenting earlier age at incidence. Aggressive tumors (Figure 2C) and young-like tumors (Figure 2D) also had peak hazard ratios early after diagnosis, with declining hazard rates thereafter. ‘Less aggressive’ SEER tumors and older-like tumors do not show this pattern. In sum, the patterns of age at incidence and hazard rate over time for young-like breast tumors are very similar to patterns presented for aggressive breast cancers based on SEER data (3).
Figure 2. Age-at-incidence distribution and hazard rate over time are similar for aggressive tumors in Surveillance Epidemiology and End Results and young-like tumors in public tumor gene expression data.
(A) Grade was used to stratify ‘aggressive’ (poorly differentiated, grade 3) tumors and ‘less aggressive’ tumors, with a left shift in the age distribution for ‘aggressive tumors. (B) The young-like tumors mirror the left shift seen with aggressive tumors, providing a biological link between age and tumor aggressiveness. (C) Aggressive tumors have a unique hazard function in SEER data, with an early peak in hazard rate (2-5 years depending on tumor characteristic modeled) followed by a decreasing hazard rate, while less aggressive tumors (grade 1 or 2 in this example), have linearly increasing hazard rate with years following diagnosis. (D) Similarly, young-like tumors have a peak hazard early (prior to 5 years following diagnosis). Tumors with older-like gene expression show the characteristic linear increase in hazard rate over time. These hazard rate curves show that young-like tumors represent more aggressive breast cancer.
Young-like tumors were more likely to have clinically aggressive characteristics (Table 3), with statistically significant associations in the largest of the three datasets (NKI): ER negative (p=0.02), high grade (p=0.005), larger (p=0.04). Substantial, though non-significant, associations in the same direction (more aggressive tumors given young-like gene expression class) were observed for Naderi et al. and UNC 337 datasets. Considering the combined dataset, significant associations held with numerous clinically aggressive phenotypes (large size, high grade, and young age). The strongest association was for high tumor grade. Young-like tumors were also more prevalent among young women in all datasets except for the Naderi et al., where the young-like signature did not correlate with patient age. However, Naderi et al. (n=52) had an older patient population (mean age of 47; compared with UNC and NKI, both mean age of 44). These results document that the normal biology of younger women is reflected inaggressive tumors that are more common in this age group.
Table 3.
Characteristics of young (≤55 years old) breast cancer patients according to young-like and old-like signature class, public microarray datasets from NKI, Naderi et al., and UNC.
| NKI | Naderi et al. | UNC 337 | Combined | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Young-like (N) | Old-like (N) | p-value | Young-like (N) | Old-like (N) | p-value | Young-like (N) | Old-like (N) | p-value | Young-like (N) | Old-like (N) | p-value | |
| ER status | ||||||||||||
| Positive | 106 | 120 | 17 | 15 | 29 | 27 | 150 | 164 | ||||
| Negative | 44 | 25 | 0.02 | 8 | 12 | 0.52 | 31 | 23 | 0.69 | 87 | 56 | 0.07 |
| Missing | 0 | 2 | 0 | 2 | ||||||||
| Size | ||||||||||||
| < 2 cm | 49 | 65 | 15 | 17 | 12 | 15 | 78 | 95 | ||||
| >= 2 cm | 101 | 80 | 0.04 | 10 | 10 | 0.95 | 48 | 36 | 0.35 | 159 | 126 | 0.02 |
| Missing | 0 | 1 | 0 | 1 | ||||||||
| Tumor Grade | ||||||||||||
| Well differentiated/1 | 31 | 44 | 9 | 5 | 2 | 5 | 42 | 54 | ||||
| Intermediate/2 | 45 | 56 | 5 | 13 | 20 | 10 | 70 | 79 | ||||
| Poorly differentiated/3 | 74 | 45 | 5.0 e-3 | 11 | 9 | 0.09 | 37 | 30 | 0.16 | 122 | 84 | 0.01 |
| Missing | 1 | 7 | 1 | 7 | ||||||||
| Subtype | ||||||||||||
| Basal | 31 | 15 | 4 | 3 | 22 | 14 | 57 | 32 | ||||
| ERBB2 | 20 | 29 | 4 | 7 | 12 | 7 | 36 | 43 | ||||
| Luminal A | 42 | 44 | 10 | 13 | 16 | 16 | 68 | 73 | ||||
| Luminal B | 44 | 37 | 4 | 3 | 9 | 11 | 57 | 51 | ||||
| Normal | 13 | 20 | 0.05 | 3 | 1 | 0.66 | 1 | 4 | 0.34 | 17 | 25 | 0.05 |
| Age | ||||||||||||
| < 30 | 1 | 3 | 0 | 0 | 3 | 2 | 4 | 5 | ||||
| 30-39 | 35 | 24 | 2 | 2 | 13 | 10 | 50 | 36 | ||||
| 40-49 | 102 | 81 | 13 | 15 | 31 | 25 | 146 | 121 | ||||
| 50-55 | 12 | 37 | 1.2 e-4 | 10 | 10 | 1 | 13 | 15 | 0.87 | 35 | 62 | 6.5e-3 |
Evaluation of correlations with previous age-related signatures
A previous study identified common signatures of aging across multiple tissues (including heart, lung, brain, muscle, and others, but not breast) and species (24). As shown in Table 4, young-like samples based on this de Magelhaes signature were younger chronologically and more likely to be young-like according to our signature. Previous studies of aging human breast were not available for comparison, but a tumor-based signature that evaluated age-related gene expression among ER-positive breast tumors (5), avoiding some of the problems of confounding by tumor subtype as described in Anders et al. (7), was evaluated. The Yau et al. signature was significantly associated with our young-like signature, but was not associated with RM patient age. The weaker correlation with age for this signature may reflect the evolution and divergence of tumor biology from normal age-related biology.
Table 4.
Association between published age signatures based classification (columns), chronological age, and reduction mammoplasty age signature class.
Conclusions
Age-associated changes in normal breast gene expression have not been reported previously. This is a gap in the literature given that age-associated gene expression has been reported for human fibroblasts and lymphocytes (29, 30) as well as brain (31), kidney (32), and skeletal muscle (33, 34). A recent meta-analysis compared aging-related changes across species and tissues, but without inclusion of mammary gland (24). Histologically, there are important compositional and morphometric changes in breast tissue with aging (35). Prior to menopause, a decline in ovarian function causes regression of both epithelium and stroma from the third to sixth decades of life, independent of previous reproductive history (36). Aging as a process spanning decades is reflected in our observation of progressive changes, and the observation that menopause was not strongly associated with changes in breast gene expression.
In our data, age-associated gene expression was functionally linked with ‘aging’ gene expression categories, but also included individual genes of interest. CDKN2A (p16) is an established biomarker and effector of mammalian aging (26), its upregulation being accompanied by shortened telomeres. As expected from the previous studies (37), we found increasing expression of p16 and decreasing expression of TERT with age. Transcripts of the gene coding tumor suppressor p53 (TP53) also changed, with p53 expression declining in older patients. Studies of p53 and aging have emphasized mouse models of breast cancer (28), but in human studies, p53 mutations are more common in younger women and basal-like tumors that occur more frequently in the young (38). These observations raise the hypothesis that increased mutation frequency in young women may reflect greater activity of p53 in young women and a resulting pressure to inactivate p53.
Other interesting developmentally-regulated pathways were also altered in aging breast. The JAK2-associated hormone-like cytokine signaling, Wnt/β-catenin signaling, and Ephrin Receptor signaling were differentially expressed with age. The hormone-dependent JAK2 signaling alterations (including higher expression of STAT5A), may reflect changes in ovarian function/estrogen signaling over time; this pathway regulating mammary gland development is responsive to estrogen and progesterone (39). The latter two pathways (Wnt/β-catenin and Ephrin receptor signaling) are known to be involved in maintaining stem cell dynamics in cancer (40, 41), but their specific roles in normal breast are relatively unexplored. Given that mammary progenitor cells are a rare cell population, these signals are unlikely to be derived specifically from stem cell populations, but may reflect the role of these pathways (and cross talk between them) in tissue architecture or cellular differentiation [reviewed in (42) and (43), for Ephrins and Wnt pathway, respectively]. We hypothesize that the changes we observe reflect alterations in survival and proliferation potential of the normal cell types that are susceptible to carcinogenesis. Alteration of these signals and normal tissue homeostasis with age may dictate pathways to malignancy and determine the aggressiveness of resulting tumors. In fact, a recent commentary emphasized the importance of altered homeostasis in age-dependent cancer rates, countering the previous notion that oncogenic mutation rates alone limit carcinogenesis at young ages (44).
Our study also identified trends that are relevant for the epidemiology of breast cancer and aging. Previous analyses of epidemiologic data have used breast cancer incidence data to draw inferences about aging of breast tissue (45, 46). These papers have suggested that the rate of aging is most rapid in the early years after menarche and before the first pregnancy, decreases with each subsequent birth, and decreases further with menopause. Use of very large datasets and anchoring of changes to particular reproductive events allowed for observation of these trends in spite of substantial inter-individual variability. While aging-related changes in undiseased tissue are a more direct route to studying aging in tissue, these studies are underpowered to dissect the composite and interactive effects of multiple demographic and exposure variables. However, it is clear in Figure 1 that there is substantial heterogeneity in the population. Premature expression of signatures reflective of older biology might predict increased risk of breast cancer. Given that a larger number of epidemiologic studies are now collecting histologically normal tissue from both diseased and unaffected individuals, development of novel risk biomarkers from normal tissue may be possible. Indeed recent studies with peripheral blood T-lymphocytes have demonstrated that molecular markers of aging do show associations with health behaviors such as physical activity and smoking status (26), suggesting that biomarkers of aging may lead to understanding of etiology.
If gene expression microenvironments in younger tissue apply selective pressures or create optimal conditions for specific subtypes, then (1) these subtypes would be more common in younger women, and (2) the tumors would be expected to differentially express the pathways common to young breast tissue. Regarding the first point, several papers have documented that different subtypes of tumors are more prevalent in younger women (e.g. ER-negative and basal-like cancer) (3, 4), which is echoed in our data showing that tumors with young-like signatures had aggressive clinical features. Regarding the second point, our work illustrates that the young-like tumors have distinct age at incidence patterns and hazard rates over time, similar to the patterns produced by stratifying on aggressive clinical characteristics. Thus our work provides a strong biological link between aging processes and the etiology of aggressive breast cancer subtypes.
While links between age-associated gene expression and epidemiologic age at incidence patterns are informative, there are caveats to our analysis. First, we used public microarray datasets to evaluate our age-associated signature classes in comparison with high grade/aggressive tumors in SEER data. While the data convincingly recapitulate the SEER patterns for age-at-incidence and hazard rate over time, it must be noted that these public datasets are not population-based samples and therefore may have substantial distributional differences from SEER in both age and tumor characteristics. While we restricted the age-range of tumors in our analysis to improve the comparability across datasets, the lack of a population-based tumor gene expression data for evaluating age-dependent signatures limits our comparability with SEER data. Second, all of the microarray datasets used were moderately sized. Therefore, it was impossible to stratify on relevant demographic variables such as race, and we were unable to detect weaker changes in gene expression with age. However, these analyses are an important first step toward characterizing some of the strongest changes induced in aging breast. Third and finally, our RM tissues were not microdissected and thus we identified changes that are common to multiple cell types. However, many aging changes may be highly stereotyped across tissue and cell types and highly conserved across organisms, given that our signature correlated with a signature derived from multiple species and multiple tissues, both stroma and epithelium rich (24).
Continued and future research may consider whether other breast cancer risk factors perturb particular breast-cancer related pathways in normal tissue. For example, if particular pathways are altered in normal breast according to body mass index (16) or parity (47), epidemiologic studies could assay these pathways in tumors and stratify cases according to whether they express these pathways. This would help to delineate the distinct causal paths that contribute to heterogeneous breast cancers. Case-only studies can identify etiologic heterogeneity with respect to particular pathways (48), and more recently, concordance of phenotypes between first and second primary cancers may help establish etiologic distinctiveness (49). Our work demonstrates that evaluating pathways in both normal tissue and in tumors can also advance our understanding of etiologic distinctiveness.
Supplementary Material
Acknowledgments
The authors acknowledge Maureen Lahti at Baystate Medical Center for contributions to patient data collection.
Grant Support: This project was supported by the National Cancer Institute and National Institutes of Environmental Health Sciences (U01-ES019472, R01-CA138255, R01-ES017400, U01-ES019548, U01-ES019466, P50CA058223), Avon Foundation, and the University Cancer Research Fund at the University of North Carolina.
Footnotes
Conflict of interest statement: None declared
The authors declare no conflicts of interests in relation to this manuscript.
References
- 1.Edwards BK, Howe HL, Ries LA, Thun MJ, Rosenberg HM, Yancik R, et al. Annual report to the nation on the status of cancer, 1973-1999, featuring implications of age and aging on U.S. cancer burden. Cancer. 2002;94:2766–92. doi: 10.1002/cncr.10593. [DOI] [PubMed] [Google Scholar]
- 2.Benz CC, Yau C. Ageing, oxidative stress and cancer: paradigms in parallax. Nat Rev Cancer. 2008;8:875–9. doi: 10.1038/nrc2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson WF, Jatoi I, Devesa SS. Distinct breast cancer incidence and prognostic patterns in the NCI's SEER program: suggesting a possible link between etiology and outcome. Breast Cancer Res Treat. 2005;90:127–37. doi: 10.1007/s10549-004-3777-3. [DOI] [PubMed] [Google Scholar]
- 4.Anderson WF, Pfeiffer RM, Dores GM, Sherman ME. Comparison of age distribution patterns for different histopathologic types of breast carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:1899–905. doi: 10.1158/1055-9965.EPI-06-0191. [DOI] [PubMed] [Google Scholar]
- 5.Yau C, Fedele V, Roydasgupta R, Fridlyand J, Hubbard A, Gray JW, et al. Aging impacts transcriptomes but not genomes of hormone-dependent breast cancers. Breast Cancer Res. 2007;9:R59. doi: 10.1186/bcr1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anders CK, Hsu DS, Broadwater G, Acharya CR, Foekens JA, Zhang Y, et al. Young age at diagnosis correlates with worse prognosis and defines a subset of breast cancers with shared patterns of gene expression. J Clin Oncol. 2008;26:3324–30. doi: 10.1200/JCO.2007.14.2471. [DOI] [PubMed] [Google Scholar]
- 7.Anders CK, Fan C, Parker JS, Carey LA, Blackwell KL, Klauber-DeMore N, et al. Breast carcinomas arising at a young age: unique biology or a surrogate for aggressive intrinsic subtypes? J Clin Oncol. 2011;29:e18–20. doi: 10.1200/JCO.2010.28.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen HJ. The cancer aging interface: a research agenda. J Clin Oncol. 2007;25:1945–8. doi: 10.1200/JCO.2007.10.6807. [DOI] [PubMed] [Google Scholar]
- 9.Benz CC. Impact of aging on the biology of breast cancer. Crit Rev Oncol Hematol. 2008;66:65–74. doi: 10.1016/j.critrevonc.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–35. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 12.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Milanese TR, Hartmann LC, Sellers TA, Frost MH, Vierkant RA, Maloney SD, et al. Age-related lobular involution and risk of breast cancer. J Natl Cancer Inst. 2006;98:1600–7. doi: 10.1093/jnci/djj439. [DOI] [PubMed] [Google Scholar]
- 14.Gatenby RA, Gillies RJ, Brown JS. Evolutionary dynamics of cancer prevention. Nat Rev Cancer. 2010;10:526–7. doi: 10.1038/nrc2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–9. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X, Casbas-Hernandez P, Bigelow C, Makowski L, Joseph Jerry D, Smith Schneider S, et al. Normal breast tissue of obese women is enriched for macrophage markers and macrophage-associated gene expression. Breast Cancer Res Treat. 2012;131:1003–12. doi: 10.1007/s10549-011-1789-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 18.Anders C, Carey LA. Understanding and treating triple-negative breast cancer. Oncology (Williston Park) 2008;22:1233–9. discussion 9-40, 43. [PMC free article] [PubMed] [Google Scholar]
- 19.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 20.Naderi A, Teschendorff AE, Barbosa-Morais NL, Pinder SE, Green AR, Powe DG, et al. A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene. 2007;26:1507–16. doi: 10.1038/sj.onc.1209920. [DOI] [PubMed] [Google Scholar]
- 21.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG, et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26:4078–85. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Cancer Institute SRP Surveillance Systems Branch. [ASCII file: yr1973_2009.seer9], Research Data (1973-2009), released April 2012, based on the November 2011 submission. 2012; Available from: www.seer.gov.
- 24.de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–81. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–6. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 26.Tsygankov D, Liu Y, Sanoff HK, Sharpless NE, Elston TC. A quantitative model for age-dependent expression of the p16INK4a tumor suppressor. Proc Natl Acad Sci U S A. 2009;106:16562–7. doi: 10.1073/pnas.0904405106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bond JA, Wyllie FS, Wynford-Thomas D. Escape from senescence in human diploid fibroblasts induced directly by mutant p53. Oncogene. 1994;9:1885–9. [PubMed] [Google Scholar]
- 28.Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci U S A. 2007;104:16633–8. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geigl JB, Langer S, Barwisch S, Pfleghaar K, Lederer G, Speicher MR. Analysis of gene expression patterns and chromosomal changes associated with aging. Cancer Res. 2004;64:8550–7. doi: 10.1158/0008-5472.CAN-04-2151. [DOI] [PubMed] [Google Scholar]
- 30.Tan Q, Zhao J, Li S, Christiansen L, Kruse TA, Christensen K. Differential and correlation analyses of microarray gene expression data in the CEPH Utah families. Genomics. 2008 doi: 10.1016/j.ygeno.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–91. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 32.Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welle S, Brooks AI, Delehanty JM, Needler N, Bhatt K, Shah B, et al. Skeletal muscle gene expression profiles in 20-29 year old and 65-71 year old women. Exp Gerontol. 2004;39:369–77. doi: 10.1016/j.exger.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Welle S, Brooks AI, Delehanty JM, Needler N, Thornton CA. Gene expression profile of aging in human muscle. Physiol Genomics. 2003;14:149–59. doi: 10.1152/physiolgenomics.00049.2003. [DOI] [PubMed] [Google Scholar]
- 35.Walker RA, Martin CV. The aged breast. J Pathol. 2007;211:232–40. doi: 10.1002/path.2079. [DOI] [PubMed] [Google Scholar]
- 36.Hutson SW, Cowen PN, Bird CC. Morphometric studies of age related changes in normal human breast and their significance for evolution of mammary cancer. J Clin Pathol. 1985;38:281–7. doi: 10.1136/jcp.38.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bazarov AV, Van Sluis M, Hines WC, Bassett E, Beliveau A, Campeau E, et al. p16(INK4a) -mediated suppression of telomerase in normal and malignant human breast cells. Aging Cell. 2010;9:736–46. doi: 10.1111/j.1474-9726.2010.00599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 39.Santos SJ, Haslam SZ, Conrad SE. Estrogen and progesterone are critical regulators of Stat5a expression in the mouse mammary gland. Endocrinology. 2008;149:329–38. doi: 10.1210/en.2007-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Incassati A, Chandramouli A, Eelkema R, Cowin P. Key signaling nodes in mammary gland development and cancer: beta-catenin. Breast Cancer Res. 2010;12:213. doi: 10.1186/bcr2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Genander M, Frisen J. Ephrins and Eph receptors in stem cells and cancer. Curr Opin Cell Biol. 2010;22:611–6. doi: 10.1016/j.ceb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15:117–34. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andres AC, Ziemiecki A. Eph and ephrin signaling in mammary gland morphogenesis and cancer. J Mammary Gland Biol Neoplasia. 2003;8:475–85. doi: 10.1023/B:JOMG.0000017433.83226.22. [DOI] [PubMed] [Google Scholar]
- 44.Degregori J. Challenging the axiom: does the occurrence of oncogenic mutations truly limit cancer development with age? Oncogene. 2012 doi: 10.1038/onc.2012.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pike MC, Krailo MD, Henderson BE, Casagrande JT, Hoel DG. ‘Hormonal’ risk factors, ‘breast tissue age’ and the age-incidence of breast cancer. Nature. 1983;303:767–70. doi: 10.1038/303767a0. [DOI] [PubMed] [Google Scholar]
- 46.Rosner B, Colditz GA. Nurses' health study: log-incidence mathematical model of breast cancer incidence. J Natl Cancer Inst. 1996;88:359–64. doi: 10.1093/jnci/88.6.359. [DOI] [PubMed] [Google Scholar]
- 47.Belitskaya-Levy I, Zeleniuch-Jacquotte A, Russo J, Russo IH, Bordas P, Ahman J, et al. Characterization of a genomic signature of pregnancy identified in the breast. Cancer Prev Res (Phila) 2011;4:1457–64. doi: 10.1158/1940-6207.CAPR-11-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Begg CB, Zhang ZF. Statistical analysis of molecular epidemiology studies employing case-series. Cancer Epidemiol Biomarkers Prev. 1994;3:173–5. [PubMed] [Google Scholar]
- 49.Begg CB. A strategy for distinguishing optimal cancer subtypes. Int J Cancer. 2011;129:931–7. doi: 10.1002/ijc.25714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.