Abstract
An X-ray crystal structure of CYP2B4 in complex with the drug paroxetine [(3S,4R)-3-[(2H-1,3-benzodioxol-5-yloxy)methyl]-4-(4-fluorophenyl)piperidine] was solved at 2.14 Å resolution. The structure revealed a conformation intermediate to that of the recently solved complex with amlodipine and that of the more compact complex with 4-(4-chlorophenyl)imidazole in terms of the placement of the F-G cassette. Moreover, comparison of the new structure with 15 previously solved structures of CYP2B4 revealed some new insights into the determinants of active-site size and shape. The 2B4-paroxetine structure is nearly superimposable on a previously solved closed structure in a ligand-free state. Despite the overall conformational similarity among multiple closed structures, the active-site cavity volume of the paroxetine complex is enlarged. Further analysis of the accessible space and binding pocket near the heme reveals a new subchamber that resulted from the movement of secondary structural elements and rearrangements of active-site side chains. Overall, the results from the comparison of all 16 structures of CYP2B4 demonstrate a cluster of protein conformations that were observed in the presence or absence of various ligands.
Introduction
Cytochromes P450 (P450) constitute a superfamily of mixed-function oxidases that metabolize a broad range of substrates, including steroids, drugs, and environmental toxicants (Johnson and Stout, 2005). Of the 57 human P450s identified, CYP2B6 is involved in the metabolism of approximately 3 to 12% of all available drugs and is inhibited by several small molecules and clinically important pharmaceuticals (Wang and Tompkins, 2008). Over the past two decades, our laboratory has conducted extensive structure-function studies of the CYP2B subfamily to elucidate the molecular basis of ligand binding and of species and strain differences in substrate oxidation (Szklarz et al., 1996; Harlow et al., 1997; Strobel and Halpert, 1997; Domanski et al., 2001; Scott et al., 2001; Spatzenegger et al., 2001; Hernandez et al., 2006; Gay et al., 2010a). In particular, CYP2B4, a rabbit enzyme that shares 78% sequence identity with CYP2B6, has been used as a model for X-ray crystallography because of its greater solubility and stability compared with the human isoform (Gay et al., 2010a; Wilderman and Halpert, 2012).
Our laboratory has determined a total of 15 crystal structures of N-terminally truncated and modified CYP2B4 either in the presence or absence of a ligand (Gay et al., 2010a; Shah et al., 2012; Zhang et al., 2013) (Note: the first CYP2B4 X-ray crystal structure was determined using a truncated and modified protein consisting of wild-type His226). As a result of the formation of a dimer involving coordination of His226 of each monomer with the heme iron of the other monomer, subsequent biochemical and crystallography work used the mutant H226Y. In this article, 2B4 refers to CYP2B4dH (H226Y) with N-terminal truncations, C-terminal His tag, and H226Y mutation unless otherwise stated.) These structures, along with biophysical and biochemical analysis, have yielded a wealth of knowledge on the conformational flexibility of the 2B subfamily of enzymes. The first crystal structure of CYP2B4 demonstrated a distinct open conformation in the absence of any ligands (Scott et al., 2003), whereas the subsequent structural analysis of CYP2B4 in complex with the inhibitor 4-(4-chlorophenyl)imidazole (4-CPI) revealed a closed enzyme conformation (Scott et al., 2004). A similar closed enzyme conformation was also observed in CYP2B4 complexed with 1-(4-chlorophenyl)imidazole (1-CPI) (Zhao et al., 2007), the clinically relevant drugs ticlopidine and clopidogrel (Gay et al., 2010b), covalent complexes with tert-butylphenylacetylene (Gay et al., 2011) and 9-ethynylphenanthrene (Zhang et al., 2013), and closed ligand free form (Wilderman et al., 2010). The crystal structures in the presence of larger imidazole inhibitors such as 1-biphenyl-4-methyl-1H-imidazole (1-PBI) (Gay et al., 2009) and bifonazole (Zhao et al., 2006) represented intermediate and expanded conformations of the protein. Our most recent structural analysis with the drug amlodipine showed a distinct open form of the enzyme that was not observed previously (Shah et al., 2012). This structure with two molecules of amlodipine bound revealed substrate access channels in CYP2B4 and 2B6. In addition, the structures revealed how these enzymes can bind drugs as large as itraconazole with high affinity. Some of the new features observed in the amlodipine complexes were consistent with the recent structural analysis of P450cam, which showed major movement of helices B′, F, and G, the F-G loop, and smaller changes in helices C, H, and I in the presence of various ligands (Lee et al., 2011).
We present here the structure of CYP2B4 in complex with the antidepressant paroxetine [(3S,4R)-3-[(2H-1,3-benzodioxol-5-yloxy)methyl]-4-(4-fluorophenyl)piperidine] (Sherman et al., 1993), which revealed a conformation of the protein intermediate to that of the 4-CPI and amlodipine complexes. Among antidepressants, paroxetine is the most potent inhibitor of bupropion hydroxylation and is thought to have the highest probability of interference with CYP2B6 function (Hesse et al., 2000). No structure of paroxetine with its major catalyst in humans, CYP2D6, is available. In retrospect, the new structure allowed us to visualize a cluster of conformations that CYP2B4 adopts on binding ligands of various sizes, in a way similar to that observed with P450cam (Lee et al., 2011). The structural analysis further demonstrates the mobility of important secondary structural elements, in particular those involved in substrate access and binding.
Materials and Methods
Materials.
Paroxetine HCl was obtained from Sigma-Aldrich (St. Louis, MO). 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) was from Calbiochem (EMD Chemicals, San Diego, CA). CYMAL-5 (5-cyclohexyl-1-pentyl-β-D-maltoside) was obtained from Anatrace (Maumee, OH). Nickel-nitrilotriacetic acid affinity resin was from Thermo Scientific (Rockford, IL). Macroprep CM cation exchange resin was received from Bio-Rad Laboratories (Hercules, CA). Amicon ultrafiltration devices were from Millipore (Billerica, MA). The pGro7 plasmid was from Takara Bio (Shiba, Japan). Escherichia coli TOPP3 cells were from Stratagene (La Jolla, CA). Hampton Research Index Screen (Aliso Viejo, CA) was used for crystallization. 3α-Hydroxyl-7α,12α-di-(((2-(trimethylamino)ethyl)phosphoryl)ethyloxy)-cholane (FA-7/234-chol) is a custom-made facial amphiphile (Zhang et al., 2007; Lee et al., 2013). Protein figures were made using the PyMOL Molecular Graphics System (DeLano Scientific, Palo Alto, CA) (DeLano, 2002).
Protein Expression and Purification.
CYP2B4 was expressed and purified in a similar manner as described previously (Shah et al., 2012). Protein expression was carried out in Luria-Bertani broth using TOPP3 cells that had been transformed with the vector pKK2B4dH (H226Y). Terrific broth was inoculated using the overnight growth culture, and protein expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside and δ-aminolevulinic acid (A600 ∼ 0.7 at 37°C) to final concentrations of 0.5 and 1 mM in the presence of ampicillin. Protein expression continued for 68–72 hours at 30°C, after which the cells were centrifuged at 4000g. The cell pellet was resuspended in 10% of the original culture volume in buffer containing 20 mM potassium phosphate (pH 7.4 at 4°C), 20% glycerol, 10 mM 2-mercaptoethanol, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and treated with lysozyme (0.2 mg/ml) before centrifugation. The spheroplasts were then resuspended in the same buffer as already described containing 500 mM potassium phosphate and sonicated three times for 45 seconds on ice. The lysate was allowed to stir for 90 minutes at 4°C in the presence of CHAPS (0.8% w/v) before ultracentrifugation for 45 minutes at 245,000g in an Optima L-80 XP ultracentrifuge using a Ti 50.2 rotor (Beckman Coulter, Fullerton, CA).
The concentration of P450 in the supernatant was determined using the reduced CO difference spectrum (Omura and Sato, 1964). The hemoprotein was purified using nickel affinity chromatography. The resin was washed with buffer containing 100 mM potassium phosphate (pH 7.4 at 4°C), 100 mM NaCl, 20% (v/v) glycerol, 10 mM 2-mercaptoethanol, 0.5 mM PMSF, CHAPS (0.5% w/v), and 1 mM histidine, and the protein was eluted using 40 mM histidine in the same buffer as described above. The P450-containing fractions were pooled and diluted 10-fold in buffer containing 5 mM potassium phosphate (pH 7.4 at 4°C), 20% (v/v) glycerol, 1 mM EDTA, 0.2 mM dithiothreitol (DTT), 0.5 mM PMSF, and CHAPS (0.5% w/v), before loading onto a Macroprep CM cation exchange column. The column was washed using 5 mM potassium phosphate (pH 7.4 at 4°C), 20 mM NaCl, 20% (v/v) glycerol, 1 mM EDTA, and 0.2 mM DTT before eluting the protein with high-salt buffer containing 50 mM potassium phosphate (pH 7.4 at 4°C), 500 mM NaCl, 20% (v/v) glycerol, 1 mM EDTA, and 0.2 mM DTT. Protein fractions were measured for their P450 content, and those with the highest A417/A280 ratios were pooled.
Crystallization and Data Collection.
The pooled protein was diluted to a final concentration of 18 μM in 50 mM potassium phosphate (pH 7.4 at 4°C), 500 mM sucrose, 500 mM NaCl, 1 mM EDTA, and 0.2 mM DTT. Paroxetine (dissolved in acetone) was added to the protein solution to make a final concentration of 180 μM, and the complex was incubated overnight at 4°C on ice. This 2B4-paroxetine complex was concentrated until the protein concentration reached 550 μM and was supplemented with 4.8 mM CYMAL-5, 1 mM paroxetine, and 0.077% (w/v) FA-7. The sitting-drop vapor diffusion method was used to screen crystallization conditions using the Hampton Research Index Screen. Crystals of 2B4 with paroxetine were obtained at 18°C after incubating the protein in a 1:1 ratio with the precipitant solution containing 0.2 M potassium sodium tartrate tetrahydrate and 20% w/v polyethylene glycol 3350. Crystals were soaked briefly in this solution containing 20% (v/v) sucrose for cryoprotection before flash-freezing in liquid nitrogen. Crystallographic data were collected remotely at Stanford Synchrotron Radiation Lightsource beam line 11-1 (Soltis et al., 2008) using 1-degree oscillations over 240 frames and 20 seconds’ exposure using a Mar 325 CCD detector at a temperature of 100 K. Crystals of 2B4-paroxetine diffracted to 2.14 Å resolution and data were integrated using iMOSFLM (Leslie, 1999) and scaled via SCALA in CCP4 (Bailey, 1994).
Structure Determination and Refinement.
The CYP2B4-paroxetine complex was determined using the coordinates of the 2B4 ligand free closed conformation structure [Protein Data Bank (PDB) ID 3MVR] as a starting model in the molecular replacement program Phaser in CCP4. The solvent content of 61.7% suggested the presence of one molecule in the asymmetric unit as determined by the Matthews coefficient analysis. Phaser (McCoy, 2007) determined the space group as P3121, and the output model was subjected to rigid body followed by restrained refinement in REFMAC (Murshudov et al., 1997). The model-building program Crystallographic Object-Oriented Toolkit (Coot) (Emsley and Cowtan, 2004) was used to build the protein manually using Fo − Fc and 2Fo − Fc electron density maps contoured at 3σ and 1σ, respectively. The PRODRG server (Schuttelkopf and van Aalten, 2004) was used to make the library description for paroxetine. After completion of the iterative refinement process, the final model was validated by MOLPROBITY (Davis et al., 2004), which ranked the structure in the 99th percentile among structures of comparable resolution, with no Ramachandran outliers or bad bond lengths or angles. The crystal structure contained a total of 231 molecules of water, three molecules of CYMAL-5 detergent, four molecules of sulfate ion, two molecules of phosphate ions, one molecule of glycerol, and protein residues 28–491 and residue 492 as the first histidine of the C-terminal His-tag. The data collection and refinement statistics for the preceding structure are shown in Table 1. Atomic coordinates and structure factors for the CYP2B4-paroxetine complex are deposited in the PDB ID 4JLT.
TABLE 1.
Data collection and refinement statistics
| Cytochrome P450 | 2B4dH (H226Y) |
|---|---|
| Ligand | Paroxetine |
| Crystal space group | P 3121 |
| Crystal unit cell parameters | |
| a = b | 90.7 Å |
| c | 153.7 Å |
| α = β | 90° |
| γ | 120° |
| Molecules per asymmetric unit | 1 |
| Data-collection statistics (values for highest-resolution shell) | |
| Beamline | SSRL 11-1 |
| Wavelength (Å) | 0.98 |
| Resolution range (Å) | 78.6–2.14 |
| Completeness (%) | 99.6 (99.4) |
| Redundancy | 11.1 (11.1) |
| Rmerge (%) | 6.4 (44.4) |
| I/σ | 8.2 (1.7) |
| No. of unique reflections | 41,055 |
| Refinement statistics | |
| R-factor | 19.1% |
| R-free | 22.9% |
| Root-mean-square deviations | |
| Bond lengths (Å) | 0.02 |
| Bond angles (°) | 1.9 |
| Ramachandran plot | |
| Preferred (%) | 98.2 |
| Allowed (%) | 1.8 |
SSRL, Stanford Synchrotron Radiation Lightsource.
Binding Pocket Calculation and Analysis.
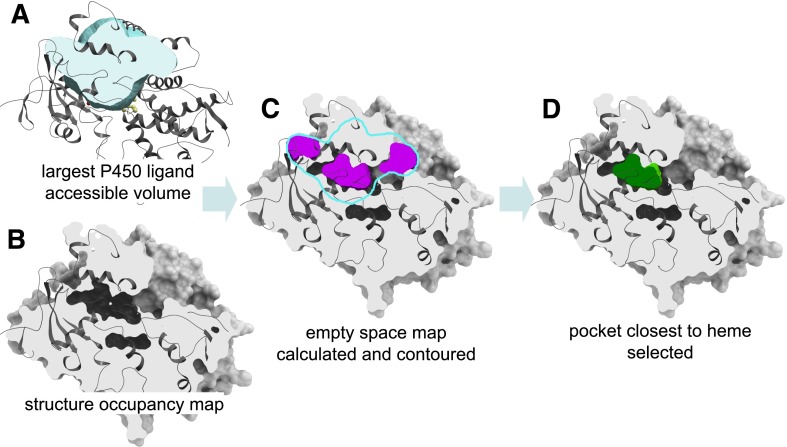
The maximal space accessible to ligands was derived as the combined Gaussian atomic density map of catalytic site ligands in all available crystallographic structures of P450 enzymes in the PDB after superimposition of the binding sites (Fig. 1A). The map was calculated on a 0.5 Å rectangular grid and flattened to avoid assigning higher value to locations occupied by more ligands. For each of the CYP2B structures in the PDB, the atomic density map was calculated in the presence of heme but after deletion of ligands and water molecules (Fig. 1B). The empty space grid map for the structure was calculated by rolling a carbon probe on a 0.5 Å grid within the maximal ligand accessible space and evaluating the probe overlap with the structure density map at each grid point. The resulting empty space map was contoured at the value of 0.45 using a map contouring algorithm implemented in ICM (Abagyan and Totrov, 1994; An et al., 2005). The algorithm produces a set of (typically discontinuous) closed meshes that approximate accurately the van der Waals surface of the protein (Fig. 1C). The mesh closest to the heme iron represents the accessible catalytic pocket space in its respective structure (Fig. 1D). Mesh volumes were calculated in ICM.
Fig. 1.
Steps of the procedure for the calculation of shapes and volumes of ligand binding pockets in P450 structures. (A) The maximal volume accessible to ligands is calculated as the combined Gaussian atomic density map of catalytic site ligands in all available crystallographic structures of P450 in the PDB after superimposition of the binding sites. (B) For each P450 structure, the atomic density map is calculated in the presence of heme and iron but after deletion of ligands and water molecules. (C) The empty space grid map for the P450 structure is calculated by rolling a carbon probe on a 0.5 Å grid within the maximal ligand accessible volume and evaluating the probe overlap with the structure density map at each grid point. The resulting empty space map is contoured at the value of 0.45, producing a set of (typically discontinuous) closed meshes closely approximating the van der Waals surface of the P450 protein. (D) The mesh closest to the heme iron atom represents the catalytic pocket in its respective structure.
Spectral Binding Titrations.
CYP2B4 spectral binding titrations were performed using paroxetine dissolved in acetone (<1% final solvent concentration) and in a buffer containing 50 mM potassium phosphate (pH 7.4 at 4°C), 500 mM NaCl, 1 mM EDTA, 0.2 mM DTT, and 500 mM sucrose. Paroxetine concentrations ranging from 0 to 5 μM were achieved by addition from a 100 µM stock added to buffer containing 1 µM P450 in a 1-ml reaction volume on an S2000 single-channel charge-coupled device rapid-scanning spectrophotometer (Ocean Optics, Inc., Dunedin, FL) using a 1-cm glass cuvette with a magnetic stirring compartment (Hellma USA, Plainview, NY) as described previously (Shah et al., 2011). Absolute spectra were recorded, and the results were fit to the equation for “tight binding” (Segel, 1975). Spectral binding constants were determined with GraphPad Prism (GraphPad Software, San Diego, CA).
IC50 Determinations.
A fluorometric assay was used to measure the rates of O-dealkylation of 7-ethoxy-(4-trifluoromethyl)coumarin (7-EFC) to 7-HFC as described previously (Hernandez et al., 2006). The recombinant proteins CYP2B4, rat NADPH-cytochrome P450 reductase (Harlow and Halpert, 1997), and rat cytochrome b5 were reconstituted at a molar ratio of 1:4:2. Each 100-µl volume in a reaction contained 50 mM HEPES, (pH 7.4 at 4°C) 15 mM MgCl2, 10 pmol P450, 40 pmol cytochrome P450 reductase, 20 pmol of cytochrome b5, 50 μM 7-EFC, and 0–100 µM paroxetine. After incubating the mixture for 5 minutes at 37°C, NADPH was added to initiate the reaction. The reaction was quenched using 20% trichloroacetic acid after 5 minutes of incubation. In addition, a control experiment in the absence of substrate was carried out, and fluorescence was measured using an F-2000 fluorescence spectrophotometer (Hitachi, Tokyo, Japan) with a λex of 410 nm and a λem of 500 nm. The IC50 values were calculated using the nonlinear regression analysis with a scientific package Igor Pro 6.1 (Wavemetrics Inc., Lake Oswego, OR).
Results
Spectral Titrations and IC50 Determination.
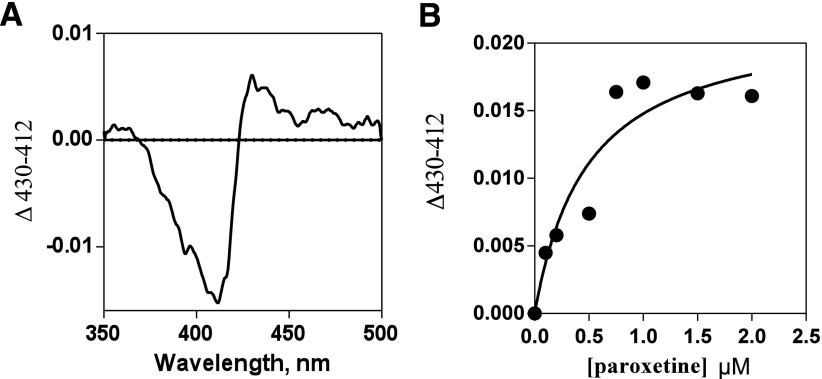
Paroxetine yielded a peak at 429 nm and a trough at 411 nm, which is consistent with type II binding of the ligand. The tight binding equation was used to determine the spectral binding constants, and a dissociation constant Kd value of 0.7 µM was obtained. The results are presented as difference spectra in Fig. 2 along with the data fitting. The IC50 value for inhibition by paroxetine of 2B4 catalyzed 7-EFC O-deethylation was 1.9 µM (unpublished data). This value is comparable to the previously reported value of 1.03 µM using wild-type CYP2B6 (Walsky et al., 2006).
Fig. 2.
Spectral titrations of CYP2B4 with paroxetine. (A) A difference spectra showing the maximum type II change obtained after the addition of paroxetine. (B) The fitting of the hyperbolic titration curve to the tight binding Eq. 2 ΔA = (ΔAmax/[E0]) - [Kd + [I0] + [E0] + (Kd + [I0] + [E0])2 - (4[E0][I0])1/2] (Segel, 1975) is shown as a solid line (B).
Structure of CYP2B4 in Complex with Paroxetine.
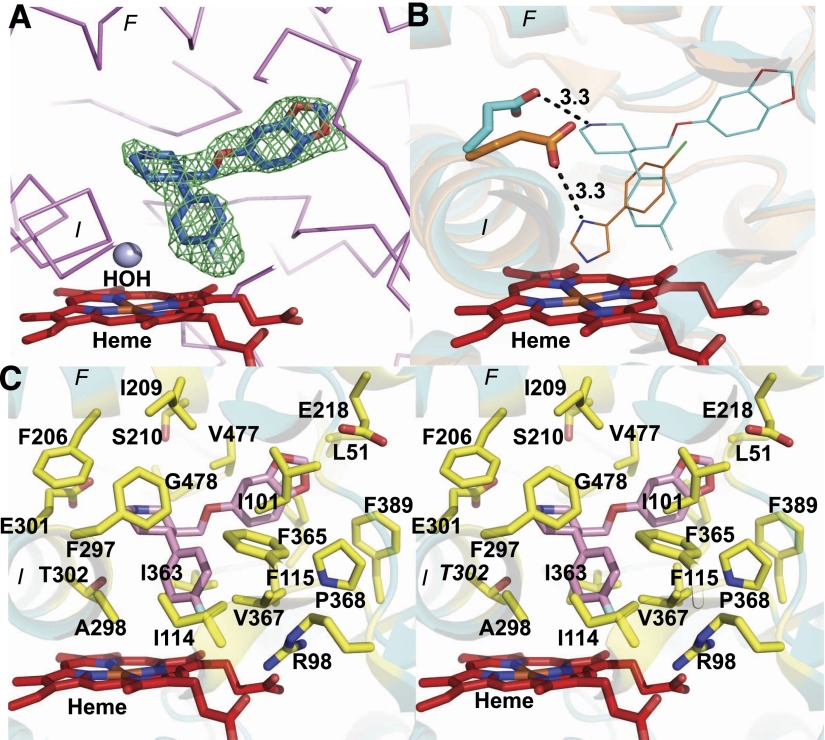
An X-ray crystal structure of CYP2B4 in complex with paroxetine was solved at 2.14 Å resolution. As shown in Fig. 3A, an unbiased electron density for the drug was observed in the active site in close proximity to the heme. Paroxetine was modeled into the density with the fluorophenyl group near heme. The ligand is circumscribed by residues I114, F115, and V367, which form a strong hydrophobic environment. The closest atom of this group of paroxetine to the heme iron is at a distance of 5 Å. Although the spectral binding data suggested nitrogen coordination to the heme iron in solution, in the structure, the piperidine nitrogen of paroxetine forms a hydrogen bond with the side-chain oxygen of E301, which might have helped in stabilizing the complex, as observed in the 2B4-4-CPI structure (Scott et al., 2004). The distance observed between the side-chain oxygen of E301 and the nitrogen atom of the respective ligand is 3.3 Å in both complexes (Fig. 3B). The methylenedioxyphenyl group of paroxetine is surrounded by the patch of residues I101, I209, F365, and F389 in the largely hydrophobic active site of CYP2B4. This region of residues circumscribes the substrate access channel 2a observed previously in the amlodipine complex of CYP2B4 (Shah et al., 2012). The residues located within a 5 Å radius of paroxetine in the active site (Fig. 3C) are L51 in the A helix; R98 in the β1 sheet; I101 in the B–B′ loop; I114 and F115 in the B-C loop; F206, I209, and S210 in the F helix; E218 in the F’ helix; F297, A298, E301, and T302 in the I helix; I363, F365, G366, V367, and P368 in the K-K′ loop; F389 in the β1-3 sheet; and G478 near the β4-2 sheet at the C-terminal end. The F206 and F297 side chains, which were previously observed to rotate to accommodate various ligands within the active site of human CYP2B6 (Shah et al., 2011), were in the same position as in the CYP2B4–4-CPI structure (Scott et al., 2004). Moreover, a CYMAL-5 detergent molecule was observed in a peripheral hydrophobic pocket near residues F202 and F296, as seen in several other structures of CYP2B4 (Shah et al., 2013). The electron density for the maltose group of one CYMAL-5 molecule located near A and A′ helices was not observed, and a third molecule of CYMAL-5 was sandwiched between the G and H helices. In addition, the density for residues 473 and 474 on the β4-1 sheet was disordered in the new 2B4 complex.
Fig. 3.
(A) An unbiased electron density Fo − Fc omit map obtained before the inclusion of paroxetine in the CYP2B4 structure at 3σ contour level, which clearly shows the presence of paroxetine (blue sticks) above the heme (red sticks). A water molecule (blue sphere) located above heme in the structure is also shown. (B) An overlay of CYP2B4-paroxetine and 4-CPI complexes showing residue E301 in each enzyme, making a hydrogen bond contact at a distance of 3.3 Å from the ligand in the respective structures. (C) Stereo view of CYP2B4 active site residues (yellow sticks) located within 5 Å radius of paroxetine (pink sticks). Heme is shown in red and the protein in cyan.
Comparison with CYP2B4–4-CPI and Amlodipine Complexes.
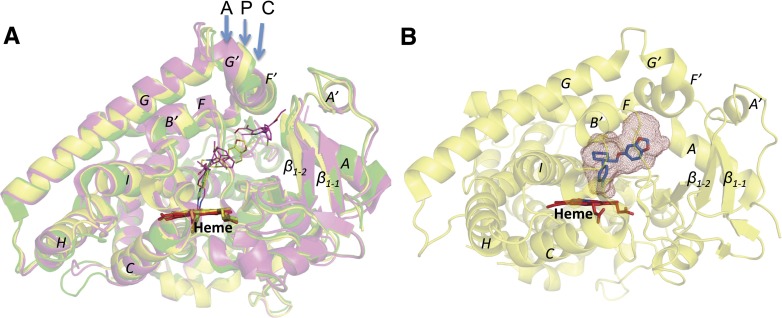
The overall alignment of three structures is shown in Fig. 4A. An overlay of the CYP2B4-paroxetine complex with the 4-CPI complex revealed an root-mean-square deviation (RMSD) of 0.36 Å. This difference results mainly from movements of several secondary structural elements that include the H-I loop region (1.5 Å), K′-K′′ loop (1.5 Å), β4 loop and β4 sheet region (1.5 Å), along with opening of the F′ and G′ region (1 Å) and the inward movement of the I-helix region from residues 296–300 (1 Å). A similar kink in the active site from residues 296–303 in the I-helix was recently observed in the 2B4 (F297A)-clopidogrel complex (Shah et al., 2013) and the previously solved 2B4-ticlopidine (Gay et al., 2010b) and 1-CPI complexes (Zhao et al., 2007). Superimposing the new structure on that of the amlodipine complex revealed noteworthy differences among several major helices and β-sheets. An overall RMSD of 0.55 was observed in a Cα overlay between the two structures. In particular, a remarkable difference was observed in the A, A′, G′, and F′ helices, which deviated by 3.5, 1.4, 1.7, and 1.6 Å, respectively. These helices represent the substrate access channel 2f that was elucidated in the 2B4-amlodipine complex with two molecules of bound ligand (Shah et al., 2012). As shown in Fig. 4A, the new CYP2B4 structure in complex with paroxetine was intermediate between that of the closed 4-CPI and more open amlodipine complexes. In addition, a difference of about 1–1.5 Å was observed between the paroxetine and amlodipine complexes in helices B′ and C, near the β1 sheets, and to some extent in the F and G helices.
Fig. 4.
(A) An overlay of CYP2B4-paroxetine (P), CYP2B4-amlodipine (A), and CYP2B4-4-CPI (C) structures is shown in yellow, magenta, and green, respectively. Ligand molecules located above heme (red sticks) are shown in thin lines for better clarity in the active site of the corresponding structures. (B) Overall structure of CYP2B4-paroxetine (yellow) with active-site cavity volume shown as red mesh. Heme and paroxetine molecules are colored in red and blue sticks, respectively.
Accessible Space and Binding Pocket in the Active Site.
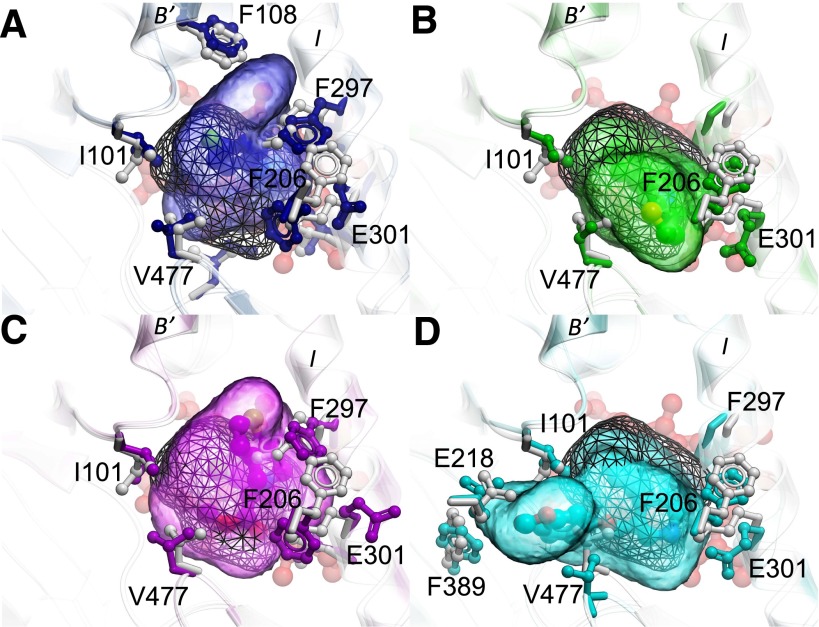
To characterize more completely the ligand-binding site in the new paroxetine bound structure, it was superimposed onto several representative CYP2B4 structures bound to 4-CPI, 1-CPI, ticlopidine, clopidogrel, or amlodipine. The active-site cavity volume of the 2B4-paroxetine complex was calculated, and the contiguous active-site cavity meshes were built. The active-site cavity volume of CYP2B4 on binding paroxetine was 557 Å3 (Fig. 4B; Table 2), which was more than twice that of the 2B4-4-CPI complex (253 Å3) and smaller than the 2B4-amlodipine complex (605 Å3) reported previously (Gay et al., 2010a; Shah et al., 2012). Two additional water molecules were added into the structure of the paroxetine complex near residues S206 and V477 to avoid what has been referred to as the “leaking effect,” where the cavity extends to the region outside of the active site (Cuff and Martin, 2004). Furthermore, the overlay in each panel of Fig. 5 demonstrates the highly variable pocket shape among the different complexes of CYP2B4. Indeed, compared with the 4-CPI bound state shown in white with gray wire pocket mesh in all panels of Fig. 5, the complexes with 1-CPI (Fig. 5A), ticlopidine (Fig. 5B), clopidogrel (Fig. 5C), and paroxetine (Fig. 5D) all feature varying degrees of side-chain and backbone movements, which open access for ligands into different subchambers in the binding site. The remarkable increase in the binding site volume in the paroxetine complex is due mainly to the movement of residues 475–480 located in the β4 sheet region. Such movement was observed previously in only the open structures of 2B4 and makes available a new subpocket adjacent to the main cavity on binding of paroxetine and similarly shaped ligands. In addition, the E301 side chain on the I-helix flips by 60 degrees away from its position in the 4-CPI complex, thus creating more volume in the active site of the new paroxetine structure. Small rearrangements of hydrophobic residues F115, F297, F365, I101, and I209 also contribute to the increased volume of the active site in the 2B4-paroxetine complex. Further movement of the preceding hydrophobic residues within the active site contributes to the even larger volume of the 2B4-amlodipine complex. In particular, F365 in the amlodipine complex of CYP2B4 flips by almost 90 degrees away from the active site, as shown previously (Shah et al., 2012).
TABLE 2.
Structural analysis of CYP2B4 with or without ligands
Active-site cavity volumes of CYP2B4-paroxetine, tert-butylphenylacetylene (tBPA) closed, L437A–4-CPI, and 9-ethynylphenanthrene (9-EP) were calculated using Voidoo (Kleywegt and Jones, 1994) as probe-occupied cavities and using a 1.4-Å probe radius. Volumes were not calculated for 2B4-open and 2B4-tBPA-open structure as a result of dimer formation that fills the active site.
| Protein | PDB | RMSD (Å) to 2B4 Paroxetine | Active-Site Cavity Volume-Å3 | Dimer | Ligand | Reference |
|---|---|---|---|---|---|---|
| 2B4-Paroxetine | 4JLT | 557 | No | Inhibitor | Current structure | |
| 2B4-Open | 1PO5 | 0.6 | Yes | No | Scott et al., 2003 | |
| 2B4-4-CPI | 1SUO | 0.36 | 253 | No | Inhibitor | Scott et al., 2004 |
| 2B4-Bifonazole | 2BDM | 1.1 | 457 | Yes | Inhibitor | Zhao et al., 2006 |
| 2B4-1-CPI | 2Q6N | 0.77 | 421 | No | Inhibitor | Zhao et al., 2007 |
| 2B4-1-PBIa | 3G93 | 1.06 | 391 | Yes | Inhibitor | Gay et al., 2009 |
| 2B4-1-PBIb | 3G5N | 1.01 | 391 | Yes | Inhibitor | Gay et al., 2009 |
| 2B4-Ligand free | 3MVR | 0.21 | 357 | No | No | Wilderman et al., 2010 |
| 2B4-Ticlopidine | 3KW4 | 0.29 | 374 | No | Substrate | Gay et al., 2010b |
| 2B4-Clopidogrel | 3ME6 | 0.39 | 416 | No | Substrate? | Gay et al., 2010b |
| 2B4-tBPA-closed | 3R1A | 0.70 | 408 | No | Covalent Inhibitor | Gay et al., 2011 |
| 2B4-tBPA-open | 3R1B | 0.88 | Yes | Covalent Inhibitor | Gay et al., 2011 | |
| 2B4-L437A-4-CPI | 3TK3 | 0.43 | 315 | No | Inhibitor | Wilderman et al., 2012 |
| 2B4-Amlodipine | 3TMZ | 0.54 | 605 | No | Inhibitor | Shah et al., 2012 |
| 2B4-F297A-Clopidogrel | 4H1N | 0.36 | 552 | No | Substrate? | Shah et al., 2013 |
| 2B4-9EP | 3UAS | 0.27 | 429 | No | Covalent Inhibitor | Zhang et al., 2013 |
Fig. 5.
An overlay of representative CYP2B4 structures with CYP2B4 (L437A) 4-CPI structure (white) as reference. The common binding pocket space accessible by ligands in all CYP2B4 structures is shown in black mesh. Heme is shown behind the binding pocket in red sticks. (A) An overlay with CYP2B4 1-CPI structure (blue) shows the extra pocket space made available by the movement of residue F297 and F108 side chains. (B and C) An overlay with the CYP2B4 ticlopidine structure (green) and clopidogrel structure (purple) shows the movement of E301 and F297 side chains, respectively, that contributes to more space in the binding pocket. (D) An overlay with the CYP2B4 paroxetine structure (blue) illustrates the movement of the loop with residues 475–480, in particular, V477 side chain, which is pushed out, thus opening access to an extra subchamber.
Discussion
The structure of CYP2B4 in complex with paroxetine presented here reveals a conformation that is intermediate to two previously solved structures, the CYP2B4-amlodipine and CYP2B4–4-CPI complexes. Comparison of the new structure with all the previously solved structures of CYP2B4 with regard to the active-site cavity volume and the overall accessible space demonstrated the movement of amino acid residue side chains that may define the pocket shape for binding of paroxetine or similar-sized ligands in the active site. The variable shape of the catalytic pocket was further illustrated in several other structures with more residue side-chain movements compared with a reference structure. Furthermore, several interesting features from the new structure were observed within the active site, including the polar contact of E301 with the nitrogen of the ligand and the lack of coordination to the heme iron.
In previous studies, coordination of the heme iron by nitrogen has been observed in several P450 structures complexed with imidazole inhibitors or other type II ligands. In solution, these compounds cause a shift of the Soret absorbance band to 421–435 (Remmer et al., 1966; Mailman et al., 1974). Such type II ligands are more often known to be inhibitors than substrates. Nonetheless, there are type II ligands that are substrates (Isoherranen et al., 2004; Pearson et al., 2011) or that coordinate heme iron via a bridging water molecule (Seward et al., 2006). Paroxetine binding to CYP2B4 yielded a difference spectrum with a trough at 411 nm and a peak at 429 nm but did not coordinate the heme iron directly or via a water molecule in the crystal structure. The closest atom of the flurophenyl group was located at a distance of 5 Å from the heme iron. In addition, unlike most type II ligands, a water molecule was located near the heme iron at a distance of 3.3 Å. Paroxetine is a substrate of CYP2D6, and it undergoes demethylenation of the methylenedioxyphenyl group to a catechol metabolite (Haddock et al., 1989). However, in the case of CYP2B4, we could not observe any metabolism of paroxetine, although a clear parent molecule was observed (personal communication, Dr. Deepak Dalvie, Pfizer, La Jolla, CA).
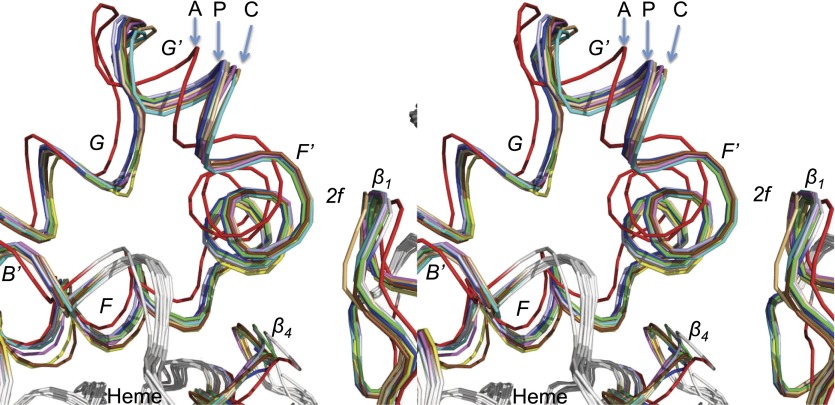
The availability of the new CYP2B4-paroxetine complex allowed us to compare and extract more detail from 15 previous CYP2B4 structures solved over the last decade. An overlay of CYP2B4-paroxetine with all these structures of CYP2B4 in the absence or presence of ligand revealed several interesting differences in overall RMSD and active site cavity volume. A total of five structures from the list in Table 2 demonstrated a wide opening of the protein as a result of dimerization. These include the first structure solved in an open conformation (PDB ID 1PO5) and subsequent 2B4 complexes with 1-PBI, bifonazole, and tert-butylphenylacetylene. The remaining structures, which were monomers, allow more sophisticated analysis of conformational changes relative to ligand size and shape. Comparison of these structures of CYP2B4 reveals many similarities with the recent analysis of a battery of P450cam structures (Lee et al., 2011). The deconvolution of 30 crystal structures of P450cam available in the PDB showed three states of the protein: open, intermediate, and closed. These correspond generally to the amlodipine, paroxetine, and 4-CPI complexes of CYP2B4, respectively. Recent Molecular Dynamics analysis (Markwick et al., 2011) suggests that larger ligands trap P450cam, in a more open conformation, whereas small ligands cause an induced fit that results in a closed conformation of the protein. A similar situation may exist with CYP2B4. As shown in Table 2, the structure of the paroxetine complex is most superimposable to the previously solved structure of CYP2B4 in a ligand-free conformation with an overall RMSD of 0.21 Å (PDB ID 3MVR). In addition, several other structures solved previously in a closed conformation showed such similarity in overall RMSD when superimposed with the CYP2B4 paroxetine complex. It is important to note that the RMSD obtained is from the overall crystal structure, which includes several flexible loops and might not be directly representative of the regions of the protein associated with ligand binding and opening of the access channels. Comparing the regions that include the F, F′, G′, G, and B′ helices and β1 sheets, where the substrate access channels were found in CYP2B4, clearly distinguishes conformational differences among these structures, as shown in Fig. 6. Additionally, differences were observed in the active site that were attributed to the movement of several secondary structural elements, such as the I-helix or the β4 loop, as well as rearrangement of the side chains of residues F206 and F297.
Fig. 6.
Stereoview of aligned Cα backbone of 11 solved CYP2B4 structures in the absence and presence of various ligands. This include structures with paroxetine (PDB ID 4JLT in blue), 4-CPI (PDB ID 1SUO in cyan), 1-CPI (PDB ID 2Q6N in brown), ligand free (PDB ID 3MVR in sky blue), ticlopidine (PDB ID 3KW4 in green), clopidogrel (PDB ID 3ME6 in magenta), tert-butylphenylacetylene (tBPA) (PDB ID 3R1A in yellow), L437A 4-CPI (PDB ID 3TK3 in yellow), amlodipine (PDB ID 3TMZ in red), F297A clopidogrel (PDB ID 4H1N in gray), and 9-ethynylphenanthrene (9-EP) (PDB ID 3UAS in light orange), as listed in Table 2. The helices F, F′, G′, G, B′, and β1 and β4 sheets involved in ligand access near channel 2f are labeled. The letters A, P, and C denote the 2B4 structures in complex with amlodipine, paroxetine, and 4-CPI, respectively.
Recent structural analysis of CYP2C9 and 2C19 demonstrated the importance of side-chain rearrangements that allow the opening of an antechamber (Reynald et al., 2012). This antechamber is a cavity situated under helix F′ and above the β1 sheet and may form a part of the substrate access channel in these CYP2C enzymes. Interestingly, F476 aligns with V477 in CYP2B4, which is displaced by up to 2 Å in the new paroxetine complex compared with all the other closed conformation structures. This movement allows the methylenedioxyphenyl group of paroxetine to set under the helix F′ facing the β1 sheet, the region of substrate access channel 2a previously identified in CYP2B4. The resulting subchamber augments the increase in overall volume of the active site compared with other closed structures. These adaptive changes within the hydrophobic active site are due mainly to the alteration of residue positions or the residue side-chain movements to accommodate ligands of various sizes. In summary, our study elucidates the structure of CYP2B4 in complex with a clinical drug and provides insights into rearrangements of secondary structural elements and residue side chains required for ligand binding. A comprehensive comparison of all the CYP2B4 structures is presented that yields knowledge of clusters of conformational states in the absence or presence of substrates and inhibitors.
Acknowledgments
The authors thank Dr. Deepak Dalvie (Pfizer, La Jolla, CA) for help with the experiments to detect CYP2B4-mediated metabolism of paroxetine and the staff at Stanford Synchrotron Radiation Lightsource, operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences, for assistance with data collection. The Stanford Synchrotron Radiation Lightsource is supported by the National Institutes of Health, the National Center for Research Resources, the Biomedical Technology Program, and the United States Department of Energy of Biological and Environmental Research.
Abbreviations
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- 1-CPI
1-(4-chlorophenyl)imidazole
- 4-CPI
4-(4-chlorophenyl)imidazole
- CYMAL-5
5-cyclohexyl-1-pentyl-β-D-maltoside
- 7-EFC
7-ethoxy-(4-trifluoromethyl)coumarin
- FA-7
3α-hydroxyl-7α,12α-di-(((2-(trimethylamino)ethyl)phosphoryl)ethyloxy)-cholane
- 7-HFC
7-hydroxy-(4-trifluoromethyl)coumarin
- P450
cytochrome P450
- paroxetine
(3S,4R)-3-[(2H-1,3-benzodioxol-5-yloxy)methyl]-4-(4-fluorophenyl)piperidine
- 1-PBI
1-biphenyl-4-methyl-1H-imidazole
- DTT
dithiothreitol
- PMSF
phenylmethylsulfonyl fluoride
- PDB
Protein Data Bank
- RMSD
root-mean-square deviation
- tBPA
tert-butylphenylacetylene
Authorship Contributions
Participated in research design: Shah, Pascual, Halpert.
Conducted experiments: Shah, Pascual, Kufareva.
Contributed new reagents or analytic tools: Zhang.
Performed data analysis: Shah, Kufareva, Pascual, Stout.
Wrote or contributed to the writing of the manuscript: Shah, Kufareva, Halpert.
Footnotes
This research was supported by the National Institutes of Health [Grants ES003619, GM098538, GM071872].
References
- Abagyan R, Totrov M. (1994) Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol 235:983–1002 [DOI] [PubMed] [Google Scholar]
- An J, Totrov M, Abagyan R. (2005) Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol Cell Proteomics 4:752–761 [DOI] [PubMed] [Google Scholar]
- Bailey S, Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763 [DOI] [PubMed] [Google Scholar]
- Cuff AL, Martin AC. (2004) Analysis of void volumes in proteins and application to stability of the p53 tumour suppressor protein. J Mol Biol 344:1199–1209 [DOI] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. (2004) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32 (Web Server issue):W615-W619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System, MacPyMOL ed., DeLano Scientific, Palo Alto, CA [Google Scholar]
- Domanski TL, He YQ, Scott EE, Wang Q, Halpert JR. (2001) The role of cytochrome 2B1 substrate recognition site residues 115, 294, 297, 298, and 362 in the oxidation of steroids and 7-alkoxycoumarins. Arch Biochem Biophys 394:21–28 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- Gay SC, Roberts AG, Halpert JR. (2010a) Structural features of cytochromes P450 and ligands that affect drug metabolism as revealed by X-ray crystallography and NMR. Future Med Chem 2:1451–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay SC, Roberts AG, Maekawa K, Talakad JC, Hong WX, Zhang Q, Stout CD, Halpert JR. (2010b) Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel. Biochemistry 49:8709–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay SC, Sun L, Maekawa K, Halpert JR, Stout CD. (2009) Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: ligand-induced structural response through alpha-helical repositioning. Biochemistry 48:4762–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay SC, Zhang H, Wilderman PR, Roberts AG, Liu T, Li S, Lin HL, Zhang Q, Woods VL, Jr, Stout CD, et al. (2011) Structural analysis of mammalian cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene: insight into partial enzymatic activity. Biochemistry 50:4903–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddock RE, Johnson AM, Langley PF, Nelson DR, Pope JA, Thomas DR, Woods FR. (1989) Metabolic pathway of paroxetine in animals and man and the comparative pharmacological properties of its metabolites. Acta Psychiatr Scand Suppl 350:24–26 [DOI] [PubMed] [Google Scholar]
- Harlow GR, Halpert JR. (1997) Alanine-scanning mutagenesis of a putative substrate recognition site in human cytochrome P450 3A4. Role of residues 210 and 211 in flavonoid activation and substrate specificity. J Biol Chem 272:5396–5402 [DOI] [PubMed] [Google Scholar]
- Harlow GR, He YA, Halpert JR. (1997) Functional interaction between amino-acid residues 242 and 290 in cytochromes P-450 2B1 and 2B11. Biochim Biophys Acta 1338:259–266 [DOI] [PubMed] [Google Scholar]
- Hernandez CE, Kumar S, Liu H, Halpert JR. (2006) Investigation of the role of cytochrome P450 2B4 active site residues in substrate metabolism based on crystal structures of the ligand-bound enzyme. Arch Biochem Biophys 455:61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse LM, Venkatakrishnan K, Court MH, von Moltke LL, Duan SX, Shader RI, Greenblatt DJ. (2000) CYP2B6 mediates the in vitro hydroxylation of bupropion: potential drug interactions with other antidepressants. Drug Metab Dispos 28:1176–1183 [PubMed] [Google Scholar]
- Isoherranen N, Kunze KL, Allen KE, Nelson WL, Thummel KE. (2004) Role of itraconazole metabolites in CYP3A4 inhibition. Drug Metab Dispos 32:1121–1131 [DOI] [PubMed] [Google Scholar]
- Johnson EF, Stout CD. (2005) Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun 338:331–336 [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. (1994) Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr 50:178–185 [DOI] [PubMed] [Google Scholar]
- Lee SCBB, Hong WX, Fu Y, Baker KA, Marcoux J, Robinson CV, Ward AB, Halpert JR, Stevens RC, Stout CD, et al. (2013) Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins. Proc Natl Acad Sci USA 110(13):E1203–E1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, Glazer EC, Wilson RF, Stout CD, Goodin DB. (2011) Three clusters of conformational states in p450cam reveal a multistep pathway for closing of the substrate access channel. Biochemistry 50:693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AG. (1999) Integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr 55:1696–1702 [DOI] [PubMed] [Google Scholar]
- Mailman RB, Kulkarni AP, Baker RC, Hodgson E. (1974) Cytochrome P-450 difference spectra: effect of chemical structure on type II spectra in mouse hepatic microsomes. Drug Metab Dispos 2:301–308 [PubMed] [Google Scholar]
- Markwick PR, Pierce LC, Goodin DB, McCammon JA. (2011) Adaptive accelerated molecular dynamics (Ad-AMD) revealing the molecular plasticity of P450cam. J Phys Chem Lett 2:158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63:32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255 [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. evidence for its hemoprotein Nature. J Biol Chem 239:2370–2378 [PubMed] [Google Scholar]
- Pearson J, Dahal UP, Rock D, Peng CC, Schenk JO, Joswig-Jones C, Jones JP. (2011) The kinetic mechanism for cytochrome P450 metabolism of type II binding compounds: evidence supporting direct reduction. Arch Biochem Biophys 511:69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmer H, Schenkman J, Estabrook RW, Sasame H, Gillette J, Narasimhulu S, Cooper DY, Rosenthal O. (1966) Drug interaction with hepatic microsomal cytochrome. Mol Pharmacol 2:187–190 [PubMed] [Google Scholar]
- Reynald RL, Sansen S, Stout CD, Johnson EF. (2012) Structural characterization of human cytochrome P450 2C19: active site differences between P450s 2C8, 2C9, and 2C19. J Biol Chem 287:44581–44591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüttelkopf AW, van Aalten DM. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363 [DOI] [PubMed] [Google Scholar]
- Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. (2003) An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc Natl Acad Sci USA 100:13196–13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EE, Spatzenegger M, Halpert JR. (2001) A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch Biochem Biophys 395:57–68 [DOI] [PubMed] [Google Scholar]
- Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. (2004) Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem 279:27294–27301 [DOI] [PubMed] [Google Scholar]
- Segel IH. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems, Wiley-Interscience, New York [Google Scholar]
- Seward HE, Roujeinikova A, McLean KJ, Munro AW, Leys D. (2006) Crystal structure of the Mycobacterium tuberculosis P450 CYP121-fluconazole complex reveals new azole drug-P450 binding mode. J Biol Chem 281:39437–39443 [DOI] [PubMed] [Google Scholar]
- Shah MB, Jang HH, Zhang Q, David Stout C, Halpert JR. (2013) X-ray crystal structure of the cytochrome P450 2B4 active site mutant F297A in complex with clopidogrel: insights into compensatory rearrangements of the binding pocket. Arch Biochem Biophys 530:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah MB, Pascual J, Zhang Q, Stout CD, Halpert JR. (2011) Structures of cytochrome P450 2B6 bound to 4-benzylpyridine and 4-(4-nitrobenzyl)pyridine: insight into inhibitor binding and rearrangement of active site side chains. Mol Pharmacol 80:1047–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah MB, Wilderman PR, Pascual J, Zhang Q, Stout CD, Halpert JR. (2012) Conformational adaptation of human cytochrome P450 2B6 and rabbit cytochrome P450 2B4 revealed upon binding multiple amlodipine molecules. Biochemistry 51:7225–7238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman TL, McDougle CJ, Price LH. (1993) Paroxetine: a new serotonin reuptake inhibitor for the treatment of depression. Conn Med 57:587–592 [PubMed] [Google Scholar]
- Soltis SM, Cohen AE, Deacon A, Eriksson T, González A, McPhillips S, Chui H, Dunten P, Hollenbeck M, Mathews I, et al. (2008) New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr D Biol Crystallogr 64:1210–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatzenegger M, Wang Q, He YQ, Wester MR, Johnson EF, Halpert JR. (2001) Amino acid residues critical for differential inhibition of CYP2B4, CYP2B5, and CYP2B1 by phenylimidazoles. Mol Pharmacol 59:475–484 [DOI] [PubMed] [Google Scholar]
- Strobel SM, Halpert JR. (1997) Reassessment of cytochrome P450 2B2: catalytic specificity and identification of four active site residues. Biochemistry 36:11697–11706 [DOI] [PubMed] [Google Scholar]
- Szklarz GD, He YQ, Kedzie KM, Halpert JR, Burnett VL. (1996) Elucidation of amino acid residues critical for unique activities of rabbit cytochrome P450 2B5 using hybrid enzymes and reciprocal site-directed mutagenesis with rabbit cytochrome P450 2B4. Arch Biochem Biophys 327:308–318 [DOI] [PubMed] [Google Scholar]
- Walsky RL, Astuccio AV, Obach RS. (2006) Evaluation of 227 drugs for in vitro inhibition of cytochrome P450 2B6. J Clin Pharmacol 46:1426–1438 [DOI] [PubMed] [Google Scholar]
- Wang H, Tompkins LM. (2008) CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9:598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PR, Gay SC, Jang HH, Zhang Q, Stout CD, Halpert JR. (2012) Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active-site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme. FEBS J 279:1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PR, Halpert JR. (2012) Plasticity of CYP2B enzymes: structural and solution biophysical methods. Curr Drug Metab 13:167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PR, Shah MB, Liu T, Li S, Hsu S, Roberts AG, Goodlett DR, Zhang Q, Woods VL, Jr, Stout CD, et al. (2010) Plasticity of cytochrome P450 2B4 as investigated by hydrogen-deuterium exchange mass spectrometry and X-ray crystallography. J Biol Chem 285:38602–38611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gay SC, Shah M, Foroozesh M, Liu J, Osawa Y, Zhang Q, Stout CD, Halpert JR, Hollenberg PF. (2013) Potent mechanism-based inactivation of cytochrome P450 2B4 by 9-ethynylphenanthrene: implications for allosteric modulation of cytochrome P450 catalysis. Biochemistry 52:355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ma X, Ward A, Hong WX, Jaakola VP, Stevens RC, Finn MG, Chang G. (2007) Designing facial amphiphiles for the stabilization of integral membrane proteins. Angew Chem Int Ed Engl 46:7023–7025 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sun L, Muralidhara BK, Kumar S, White MA, Stout CD, Halpert JR. (2007) Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4. Biochemistry 46:11559–11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. (2006) Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J Biol Chem 281:5973–5981 [DOI] [PubMed] [Google Scholar]