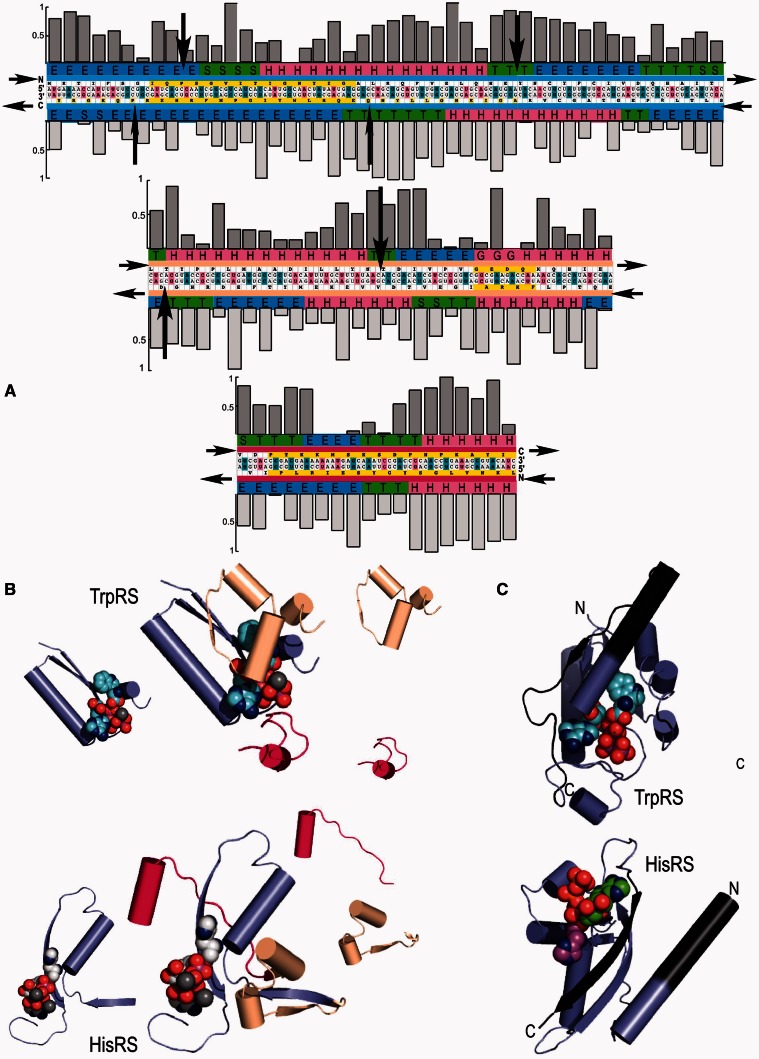
Fig. 1.
Modularity in class I and II aminoacyl-tRNA synthetase Urzymes. Complementary modularity within aminoacyl-tRNA synthetase active sites. (A) Summary of information derived from the respective TrpRS and HisRS multiple sequence alignments. The Bacillus stearothermophilus TrpRS and Escherichia coli HisRS sequences are aligned antisense to each other, consistent with the Rodin–Ohno hypothesis. The three modular fragments comprising the Urgene are indicated by a thin colored strip between sequence and secondary structure codes. Yellow blocks indicate the motifs used to anchor the three coding blocks, which were assembled end-to-end to form a single “gene.” Codon middle base pairs are magenta, unpaired middle bases are blue. Sequence entropies of class I (darker shading) and class II (lighter shading) are shown as histograms above and below the sequences. Conserved positions have low sequence entropies. Significant numbers of inserted residues in the respective MSAs occur at sites indicated by black arrows. Secondary structures from the crystal structures of the full-length enzymes are encoded as: H = α-helix, E = extended (β), T = turn, and S = loop. (B) Decomposition of tertiary structures inferred from crystal structures of the intact enzymes into the three modular coding blocks, colored as in (A). Aminoacyl-5′AMP intermediates are denoted by spheres. Note that the class I (TrpRS) active site encloses the indole moiety completely, while the imidazole moiety of histidine has extensive solvent-exposed surface area. (C) Relationship between the Urgene in (A) and the active Urzymes. Gaps between the excerpted fragments (dark gray additions) represent the remaining puzzle of reconstructing an alignment coding two intact active enzymes (see Discussion).