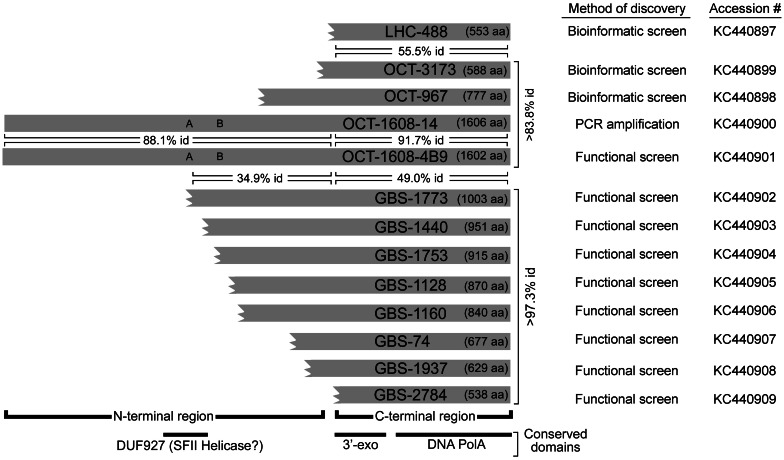
Fig. 2.
Schematic alignment of polA gene products of the indicated clones showing length in amino acids (aa) of conserved domains, method of discovery. Three letter codes specify the source hot spring: LHC (CA), OCT (YNP, WY), and GBS (NV). Truncated ORFs are shown with broken amino termini. Percent amino acid identities for the carboxy- and amino-terminal domains are indicated in horizontal brackets for selected pairs. Minimum percent identities for all pairwise comparisons within each biogeographic cluster are indicated in vertical text to the right of the sequences. A and B denote Walker A (P-loop) and B box motifs proposed to be involved in NTP binding and hydrolysis. GenBank accession numbers corresponding to the polA genes are shown.