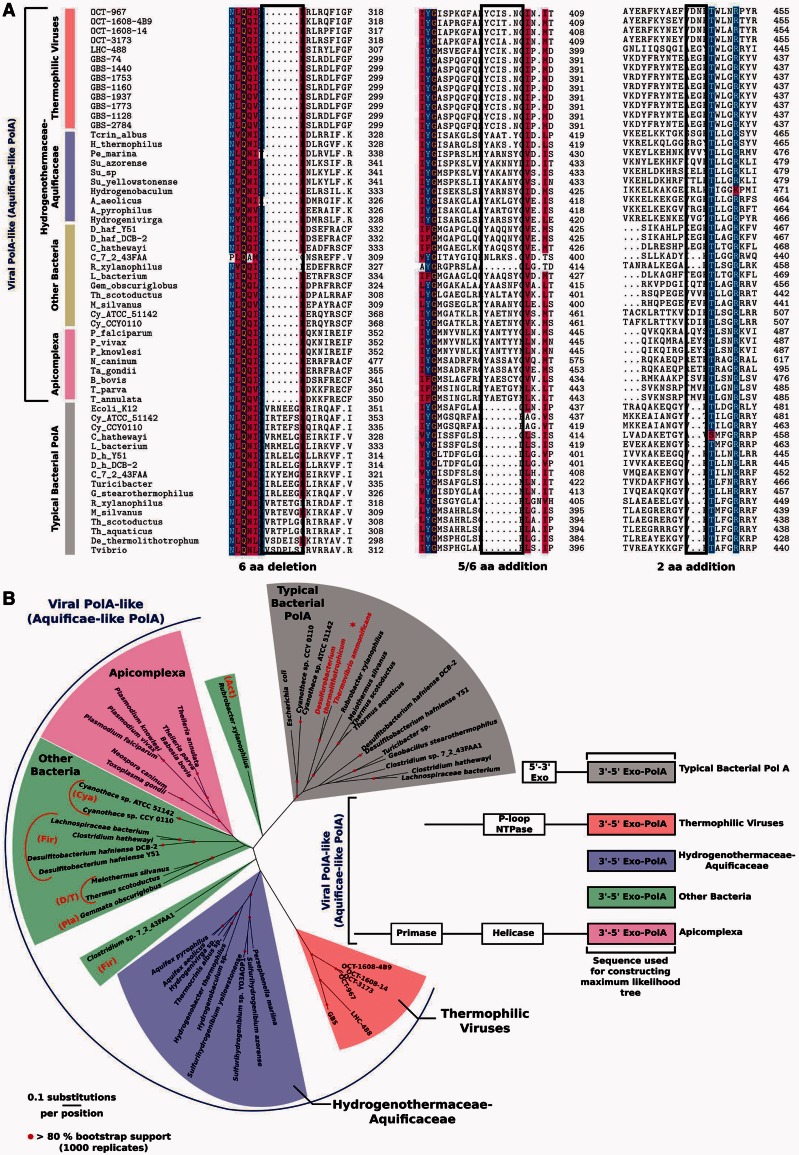
Fig. 3.
Alignment of conserved indels of viral, Aquificae, and Apicoplast polA genes based on signature motifs identified in (Griffiths and Gupta 2006) (panel A). Representative prototypical bacterial PolAs are shown at the bottom for comparison. Maximum-likelihood phylogeny of carboxy-terminal 3′exo/pol domains of viral and cellular polA-like genes (panel B). Representative prototypical bacterial PolA proteins are shown for comparison (gray) with the Aquificae family Desulfurobacteriaceae emphasized (asterisk, red text). The region used for the analysis consisted of the 588 C-term amino acids corresponding to clone OCT-3173 (fig. 1). The schematic alignment (right) highlights the carboxy terminal regions used for analysis and shows divergence of amino-terminal PolA proteins. Branches with more than 80% bootstrap values (1,000 replicates) are indicated. Phylum-level designations for non-Aquificae bacterial hosts of viral polA genes are indicated by abbreviations as follows: Act, Actinobacteria; Cya, Cyanobacteria; Fir, Firmicutes; D/T, Deinococcus-Thermus; Pla, Planctomycetes. An analysis with the same alignment and a 50% mask shows similar results (supplementary fig. S3, Supplementary Material online).