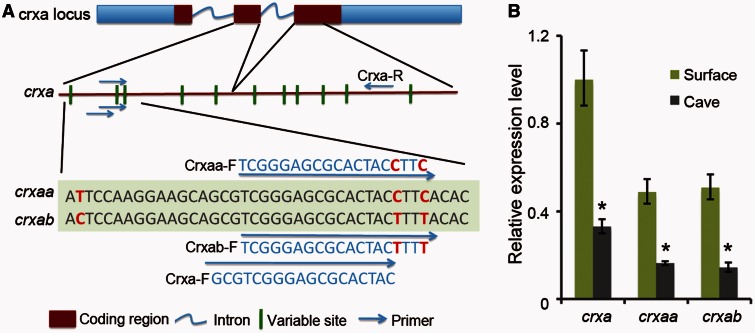
Fig. 8.
(A) Our de novo assembled Sinocyclocheilus transcriptome identified two co-orthologs, crxaa and crxab, of the single zebrafish crxa gene; these arose in the tetraploid origin of Sinocyclocheilus. The crxaa and crxab genes have several identifying nucleotide variations. To distinguish the expression level of different crxa paralogs, we designed primers to detect crxaa and crxab with real-time PCR using RNA isolated from eyes of each species. (B) The expression levels of crxa paralogs were quantified and normalized to beta-actin-1. Relative expression values are mean ± SD of at least three independent experiments. Results showed that both crxaa and crxab were downregulated in cavefish, but that there was no obvious difference between crxaa and crxab within each species.