Abstract
Binding of IP3 (inositol 1,4,5-trisphosphate) to the IP3-binding core (residues 224–604) of IP3Rs (IP3 receptors) initiates opening of these ubiquitous intracellular Ca2+ channels. The mechanisms are unresolved, but require conformational changes to pass through the suppressor domain (residues 1–223). A calmodulin-binding peptide derived from myosin light chain kinase uncouples these events. We identified a similar conserved 1-8-14 calmodulin-binding motif within the suppressor domain of IP3R1 and, using peptides and mutagenesis, we demonstrate that it is essential for IP3R activation, whether assessed by IP3-evoked Ca2+ release or patch-clamp recoding of nuclear IP3R. Mimetic peptides specifically inhibit activation of IP3R by uncoupling the IP3-binding core from the suppressor domain. Mutations of key hydrophobic residues within the endogenous 1-8-14 motif mimic the peptides. Our results show that an endogenous 1-8-14 motif mediates conformational changes that are essential for IP3R activation. The inhibitory effects of calmodulin and related proteins may result from disruption of this essential interaction.
Keywords: 1-8-14 motif; calcium signalling; calmodulin; inositol 1,4,5-trisphosphate receptor; myosin light chain kinase (MLCK)
Abbreviations: BCR, B-cell receptor; CaBP1, Ca2+-binding protein 1; CaM, calmodulin; CLM, cytosol-like medium; IP3, inositol 1,4,5-trisphosphate; IBC, IP3-binding core; IP3R, IP3 receptor; MLCK, myosin light chain kinase; NT, N-terminus; RyR, ryanodine receptor; SD, suppressor domain
INTRODUCTION
Ca2+ channels allow most electrical and many chemical signals to be transduced into the changes in cytosolic Ca2+ concentration that regulate almost every aspect of cellular activity [1]. Most Ca2+ channels are also regulated by Ca2+, either directly or via CaM (calmodulin) [2]. This provides feedback regulation of Ca2+ signalling and it allows Ca2+ channels to evoke regenerative Ca2+ signals [3]. The latter are important because they underpin the versatility of Ca2+ as an intracellular messenger, permitting it to function either locally or globally [1].
Two major families of intracellular Ca2+ channels, IP3Rs [IP3 (inositol 1,4,5-trisphosphate) receptors] and RyRs (ryanodine receptors), share many structural [4,5] and functional [5–7] properties. Most notably, all IP3Rs and RyRs are stimulated by low concentrations of cytosolic Ca2+ and inhibited by higher concentrations. Ca2+-binding sites within the RyR itself can mediate this biphasic Ca2+ regulation [7], but, for IP3Rs, it remains unclear whether additional Ca2+-binding proteins are required [6]. None of the many Ca2+-binding sites in RyRs [8] or IP3Rs [9] has been unambiguously associated with Ca2+ regulation of channel gating [10,11], although mutation of a single equivalent residue in RyRs or IP3Rs (Glu2100 in IP3R1) modulates their Ca2+-sensitivity [11]. Both families of intracellular Ca2+ channels are also regulated by CaM, a ubiquitously expressed and highly conserved Ca2+-binding protein [12]. Related proteins with EF-hand Ca2+-binding structures, such as S100A and CaBP1 (Ca2+-binding protein 1), also regulate RyRs and IP3Rs, but the physiological significance of these interactions between intracellular Ca2+ channels and CaM or related proteins is unresolved [13,14]. Despite some conflicting evidence [15], CaM seems not to be essential for Ca2+ regulation of RyRs or IP3Rs [16–18], but it does regulate both channels and it modulates their responses to Ca2+ [19–21].
All IP3Rs are inhibited by Ca2+–CaM [22], but neither of the two CaM-binding sites within IP3R1, nor a third that is created by alternative splicing [23], clearly mediates this inhibition of IP3-evoked Ca2+ release. The central site [24] (Figure 1A) mediates neither Ca2+ nor CaM regulation of IP3R activity [16,17] and it is absent from IP3R3. The functional role of the split N-terminal site (Figure 1A), one component of which may also bind CaBP1 [25], is also unclear. It has been proposed to bind CaM and thereby to inhibit IP3R activity, but only when Ca2+ has bound elsewhere [26]. The evidence that CaM inhibits IP3R only in the presence of Ca2+, without CaM itself providing the Ca2+-sensor, is persuasive [26], but there is no compelling evidence to link this to the N-terminal CaM-binding site [27].
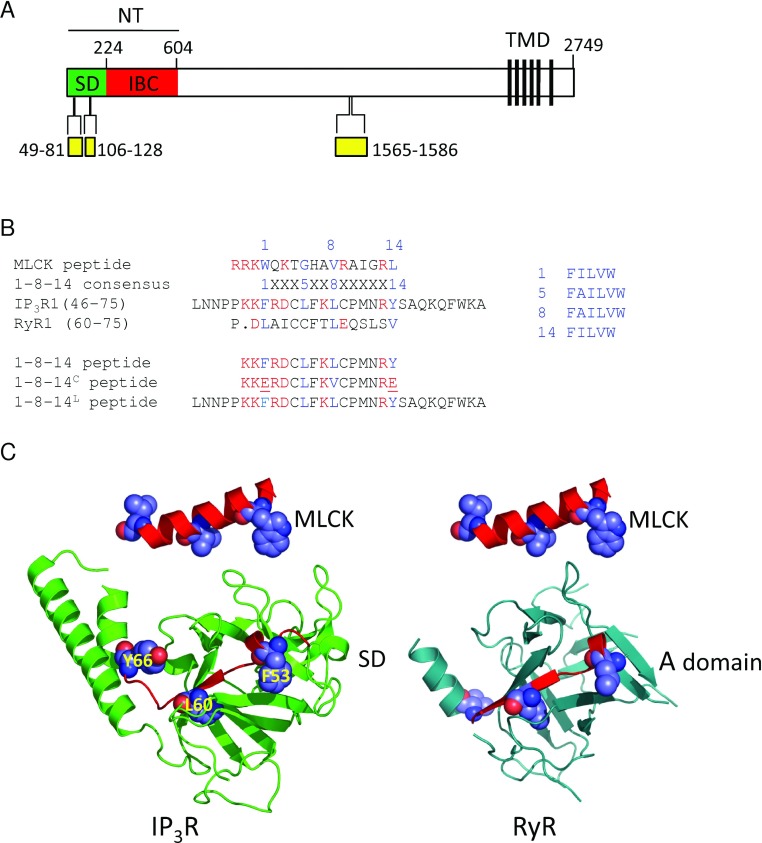
Figure 1. A putative 1-8-14 motif within the SD of the IP3R.
(A) Key features of a rat IP3R showing the NT, with its component parts (SD and IBC), the six C-terminal transmembrane domains (TMD) that form the pore and the CaM-binding domains (yellow). Residue numbers are shown. (B) Comparison of 1-8-14 motifs showing the conserved hydrophobic residues of the consensus sequence in blue. Charged residues within the 1-8-14 motif are highlighted in red because the consensus motif has a net charge of +3 to +6. The lower panel shows the peptides used with mutated residues underlined. (C) Structure of the SD of IP3R1 (PDB code 1XZZ) and the equivalent region (A domain) of RyR1 (PDB code 3HSM) with the pseudo-1-8-14 motif highlighted and compared with MLCK in the structure it adopts when bound to Ca2+–CaM (PDB code 1QTX).
The links between CaM binding and function are better understood for RyRs, although the effects differ between RyR subtypes [7]. A single site on each RyR1 subunit (residues 3614–3643 in rabbit RyR1), which is conserved in all RyRs, binds the C-terminal lobe of both apo-CaM and Ca2+–CaM and appears to mediate the functional effects of CaM [20,28,29]. As this tethered CaM binds Ca2+, it migrates towards the NT (N-terminus) of the binding site and the CaM switches from activating RyR1 to inhibiting it [19]. The CaM-binding site of RyR1 also engages other CaM-like domains, notably the C-terminus of the L-type Ca2+ channel which inhibits RyR1 activity [30], and perhaps an EF-hand-like structure within the C-terminal region of RyR1 which binds Ca2+ and modulates Ca2+ regulation of RyR [31]. These observations suggest that the CaM-binding domain of RyR also mediates important inter- and intra-molecular interactions, and that the complex effects of CaM and related proteins may, at least in part, result from disrupting these interactions [29,31,32].
For IP3Rs, IP3 binding to the IBC (IP3-binding core) (residues 224–604) (Figure 1A) initiates the conformational changes that lead to opening of a pore formed by the C-terminal transmembrane domains of each of the four IP3R subunits [5,33]. These conformational changes pass via the N-terminal SD (suppressor domain) (residues 1–223), which is essential for IP3R activation. Indeed, the major conformational changes associated with IP3R activation appear to occur within the NT (residues 1–604) [5,33]. Although both IP3 and Ca2+ are required for IP3R activation [6,34], it is not yet clear how the conformational changes initiated by IP3 lead to Ca2+ binding and then to gating of the pore. It is therefore intriguing that a CaM-binding peptide derived from MLCK (myosin light chain kinase), which comprises a 1-8-14 CaM-binding sequence [35], reversibly inhibits IP3-evoked Ca2+ release [36] via all three vertebrate IP3R subtypes. Furthermore, MLCK peptide is more potent in the presence of Ca2+ [35]. This inhibition is entirely independent of CaM and involves interaction of MLCK peptide with the NT in a manner that requires the SD [35]. We speculate, by analogy with RyRs, that inhibition of IP3Rs by MLCK peptide might result from disruption of an interaction between endogenous CaM-like and CaM-binding domains within IP3Rs, and that, for IP3Rs, this interaction is essential for activation. In the present study, we explored this hypothesis further.
EXPERIMENTAL
Materials
Cell culture materials were from Gibco, except for fetal bovine serum (Sigma). CaM purified from bovine brain was from Calbiochem. [3H]IP3 (18 Ci/mmol) was from PerkinElmer. IP3 was from Alexis Biochemicals. Peptides were synthesized and purified by Sigma or New England Peptide, and each was shown to be >90% pure by HPLC. The peptide sequences are listed in Supplementary Table S1 (at http://www.BiochemJ.org/bj/449/bj4490039add.htm).
Site-directed mutagenesis
The NT (residues 1–604) and IBC (residues 224–604) of rat IP3R1 were amplified by PCR from the full-length receptor clone lacking the SI splice region (GenBank® accession number GQ233032.1) as described previously [33]. The fragments were ligated into pTrcHis A (Invitrogen) to allow expression of N-terminally His6-tagged proteins. Mutagenesis of the 1-8-14 motif within the NT used the QuikChange® II XL site-directed mutagenesis kit (Stratagene) for single mutants (F53E, L60E, Y66E and K52E) and the QuikChange® multi-site-directed mutagenesis kit for the double mutant (F53E and Y66E). The primers used are listed in Supplementary Table S2 (at http://www.BiochemJ.org/bj/449/bj4490039add.htm). The same primers and conditions were used for mutagenesis of full-length IP3R using IP3R1 in the pENTR 1A vector. Full-length constructs were subcloned into pcDNA3.2/V5-DEST for expression in DT40 cells. The complete sequence of every mutant construct was verified by sequencing.
Culture and stable transfection of DT40 cells
DT40 cells in which the genes for all three IP3R subtypes had been disrupted (DT40-KO) [37] and DT40 cells stably expressing rat IP3R1 (DT40-IP3R1) were grown in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum, 1% (v/v) heat-inactivated chicken serum, 2 mM l-glutamine and 50 μM 2-mercaptoethanol. Cells were grown in suspension in 175 cm2 flasks at 37°C in an atmosphere of 5% CO2. They were used or passaged when they reached a density of ~2×106 cells/ml. To generate stable cell lines expressing mutant IP3R, the mutant construct in pcDNA3.2/V5-DEST was linearized, and DT40 cells were transfected by nucleofection (Amaxa, protocol B-23). Cell lines were selected with G-418 (2 mg/ml) and screened initially by Western blotting using a peptide antiserum to IP3R1 [38] as described previously [33], and then using the functional assay described below.
Ca2+ release from the intracellular stores of permeabilized cells
The free Ca2+ concentration of the intracellular stores of permeabilized cells was measured using a low-affinity Ca2+ indicator trapped within the endoplasmic reticulum as reported previously [39]. Briefly, DT40 cells (4×107 cells/ml) were suspended in HBS (Hepes-buffered saline: 135 mM NaCl, 5.9 mM KCl, 11.6 mM Hepes, 1.5 mM CaCl2, 11.5 mM glucose and 1.2 mM MgCl2, pH 7.3) containing 1 mg/ml BSA, 0.4 mg/ml Pluronic F127 and 20 μM mag-fluo-4/AM (Invitrogen). After 1 h at 20°C in the dark with gentle shaking, cells were centrifuged at 650 g for 2 min and resuspended to 107 cells/ml in Ca2+-free CLM (cytosol-like medium) (20 mM NaCl, 140 mM KCl, 1 mM EGTA, 20 mM Pipes and 2 mM MgCl2, pH 7.0) containing 20 μg/ml saponin. After incubation at 37°C with gentle shaking for 4 min, permeabilized cells were centrifuged at 650 g for 2 min and resuspended in Mg2+-free CLM, supplemented with CaCl2 to give a final free Ca2+ concentration of 220 nM. The free Ca2+ concentration of CLM was calculated using the MaxChelator program (http://maxchelator.stanford.edu) and then measured using fluo-3 or fura-2. Cells were then washed, resuspended in Mg2+-free CLM containing 10 μM FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone) to inhibit mitochondria, and distributed into a 96-well plate (106 cells in 50 μl of CLM/well). After centrifugation, fluorescence from the luminal indicator was recorded using a FlexStation II platereader (Molecular Devices) equipped to allow automated additions [39]. In all experiments, the intracellular stores were allowed to load to steady-state with Ca2+ after addition of MgATP. IP3 was then added with thapsigargin (1 μM, to inhibit Ca2+ reuptake). The Ca2+ release evoked by IP3 is expressed as a fraction of the ATP-dependent Ca2+ uptake.
Patch-clamp recording
Currents were recorded from patches excised from the outer nuclear envelope of DT40 cells expressing recombinant rat IP3R1 using symmetrical caesium methanesulfonate (140 mM) as the charge-carrier. The composition of recording solutions and methods of analysis were otherwise as described previously [40].
Expression of N-terminal fragments of IP3R
The pTrcHis constructs were used for expression of N-terminally His6-tagged proteins in Escherichia coli strain BL21(DE3) cells. Before use for [3H]IP3 binding, proteins were cleaved from the His6 tags using biotinylated thrombin (Novagen) at the engineered thrombin-cleavage site [33]. Complete cleavage was verified by Western blotting using an anti-His6 antibody. The proteins were used for [3H]IP3 binding without further purification [33].
[3H]IP3 binding
Equilibrium-competition binding assays were performed at 4°C for 5 min in CLM (500 μl) with a free Ca2+ concentration of 220 nM and containing [3H]IP3 (0.75–1.5 nM), bacterial lysate (10 μg of protein for IBC and 100 μg of protein for NT) or cerebellar membranes (50 μg of protein) and competing ligands. Non-specific binding was defined by addition of 10 μM IP3. Bound and free [3H]IP3 were separated by centrifugation at 20000 g for 5 min, after addition of poly(ethylene glycol) (15% final concentration) and γ-globulin (0.75 mg) for soluble proteins. Results were analysed by fitting to a Hill equation (using GraphPad Prism) from which the IC50 (half-maximal inhibitory concentration) and thereby the Kd (equilibrium dissociation constant) were calculated [33].
Western blotting
Cells in Ca2+-free CLM containing 2-mercaptoethanol (1 mM) and protease inhibitors were lysed by addition of PopCulture (10%), lysozyme (10 μg/ml), DNAse (5 units/ml) and RNAse (10 μg/ml). The proteins were separated using SDS/PAGE pre-cast mini-gels (Invitrogen) and transferred on to a PVDF membrane using an Iblot dry-transfer apparatus (Invitrogen). The primary antibodies were rabbit anti-His6 (1:3000 dilution) (Sigma) and anti-IP3R1 (1:1000 dilution) [33]. HRP (horseradish peroxidase)-conjugated anti-rabbit secondary antibodies (1:5000 dilution) (AbCam) and the Super Signal West Pico chemiluminescence reagent (Pierce) were used to detect immunoreactivity. Bands were quantified using GeneTools software (Syngene).
Statistical analysis
For comparisons of Kd, EC50 (half-maximally effective concentration) or IC50 values, their negative logarithms (pKd, pEC50 and pIC50; means±S.E.M.) were used for statistical analyses. For clarity, some Figures show normalized results, but all statistical analyses were performed on the raw data using paired or unpaired Student's t tests. P<0.05 was considered significant.
RESULTS AND DISCUSSION
Reversible inhibition of IP3-evoked Ca2+ release by an endogenous 1-8-14 peptide
A sequence within the SD of all known IP3Rs (residues 53–66 in rat IP3R1; Supplementary Figure S1 at http://www.BiochemJ.org/bj/449/bj4490039add.htm) includes the critical hydrophobic residues of a 1-8-14 CaM-binding motif appropriately oriented within the known structure of the SD [41] (Figures 1B and 1C) and with the required net positive charge [35]. The sequence lies within one of the two regions (residues 49–81; Figure 1A) within the NT reported to bind CaM [42] and CaBP1 [14]. A similar sequence is present within the N-terminal of all RyRs (Supplementary Figure S1). To test our hypothesis that inhibition of IP3R by MLCK peptide results from disruption of an essential interaction involving an endogenous 1-8-14 motif, we assessed the effects of a peptide derived from this motif (1-8-14 peptide; Figure 1B and Supplementary Table S1) on IP3-evoked Ca2+ release.
The 1-8-14 peptide inhibited IP3-evoked Ca2+ release via IP3R1 without affecting either Ca2+ uptake or the sensitivity (EC50) to IP3 (Figures 2A–2D). A maximally effective concentration of the peptide reduced the maximal response to IP3 by 77±7%. The IC50 for 1-8-14 peptide was 767 μM (pIC50, 3.1±0.25) (Figure 2C). Neither a mutant 1-8-14 peptide, in which two critical hydrophobic residues are mutated (1-8-14C, 3 mM) nor a scrambled peptide (1-8-14S, 3 mM) had any effect on IP3-evoked Ca2+ release (Figure 2C). Both MLCK peptide (isoelectric point, pI 14.0) and 1-8-14 peptide (pI 11.6) are very basic and might therefore have inhibited IP3-evoked Ca2+ release by binding directly to IP3. We demonstrated previously that this was not the case for MLCK peptide [35], and it is also unlikely for the 1-8-14 peptide. The 1-8-14 and 1-8-14S peptides are equally basic, but only the former inhibited IP3R; the percentage inhibition caused by 3 mM 1-8-14 peptide is similar for all IP3 concentrations (~75%), and neither was the inhibition reduced by increasing the IP3 concentration beyond that required to stimulate maximal Ca2+ release (Figure 2B). We conclude that 1-8-14 peptide inhibits IP3-evoked Ca2+ release by binding to IP3R.
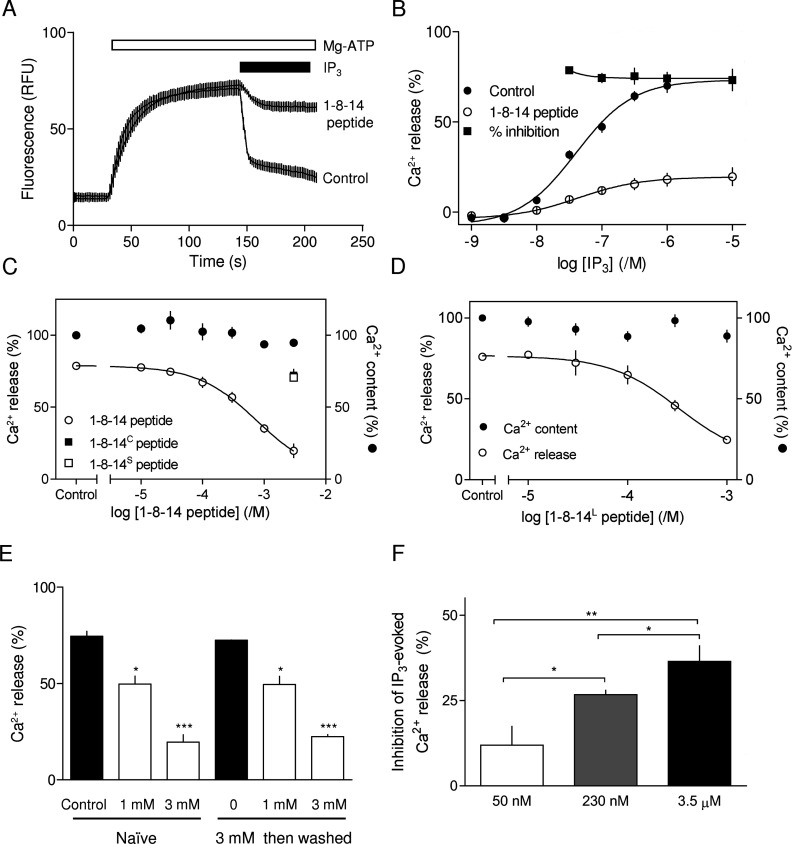
Figure 2. Inhibition of IP3R by 1-8-14 peptide.
(A) Typical recording of the free Ca2+ concentration within the endoplasmic reticulum of a population of permeabilized DT40-IP3R1 cells showing Ca2+ uptake after addition of MgATP (1.5 mM), release of Ca2+ after addition of IP3 (10 μM, with 1 μM thapsigargin to inhibit Ca2+ re-uptake) and inhibition of that release by 1-8-14 peptide (3 mM, present throughout as indicated, upper trace). Results are means±S.E.M. for three replicates from a single experiment. (B) Concentration-dependent release of intracellular Ca2+ stores by IP3 alone or after pre-incubation for 2.5 min with 1-8-14 peptide (3 mM). Inhibition by 1-8-14 peptide at each IP3 concentration is also shown (%). 1-8-14 peptide caused a significant decrease in the maximal response (P<0.001) without significantly changing the sensitivity to IP3. (C and D) Permeabilized cells pre-incubated for 10–20 min with the indicated concentrations of peptide were stimulated with IP3 (10 μM, in the continued presence of peptide). Results show the Ca2+ content of the stores before addition of IP3, and the Ca2+ release evoked by IP3. (E) Permeabilized cells were incubated alone or with 1-8-14 peptide (3 mM) for 10–20 min, washed and then resuspended in CLM. Ca2+ release by IP3 (10 μM) was then measured after a further incubation for 10–20 min with the indicated concentrations of 1-8-14 peptide. The Ca2+ release evoked by IP3 with and without peptide is shown for naive cells and after the pre-treatment with 3 mM peptide. The results establish that the effects of 1-8-14 peptide are fully reversible. (F) Permeabilized cells pre-incubated with or without 1-8-14 peptide (1 mM) for 10–20 min were stimulated with a maximally effective concentration of IP3 in the continued presence of peptide in CLM with the indicated free Ca2+ concentration. Results show the inhibition of IP3-evoked Ca2+ release (%) by 1-8-14 peptide at each free Ca2+ concentration. Results in (B)–(F) are means±S.E.M. (n≥3). *P<0.05, **P<0.01 and ***P<0.001.
The 1-8-14 peptide is only 16 residues long. A longer peptide (30 residues, 1-8-14L), which includes additional N- and C-terminal residues that are conserved in all IP3Rs (Figure 1B and Supplementary Figure S1), also inhibited IP3-evoked Ca2+ release without affecting Ca2+ uptake (Figure 2D). Although IP3R may be slightly more sensitive to the longer peptide (IC50, 326 μM; pIC50, 3.5±0.25) than to the 1-8-14 peptide (767 μM, 3.1±0.25); the difference was not statistically significant. Subsequent studies used the shorter 1-8-14 peptide because it was less expensive.
The results shown in Figure 2(E) demonstrate that the effects of a maximally effective concentration of 1-8-14 peptide (3 mM) are fully reversible. These experiments, which require extensive washing of the cells between successive challenges with the peptide, confirm that the inhibition of IP3Rs by the 1-8-14 peptide, like that by MLCK peptide [35], does not result from dissociation of CaM from IP3R [36]. Our previous study demonstrated that MLCK peptide more potently inhibited IP3R when the cytosolic free Ca2+ concentration was increased [35]. Similar results were obtained with 1-8-14 peptide (Figure 2F). We conclude that 1-8-14 peptide inhibits IP3-evoked Ca2+ release by binding to the IP3R and the inhibition is enhanced at elevated cytosolic Ca2+ concentrations.
Inhibition of single-channel currents through IP3Rs by 1-8-14 peptide
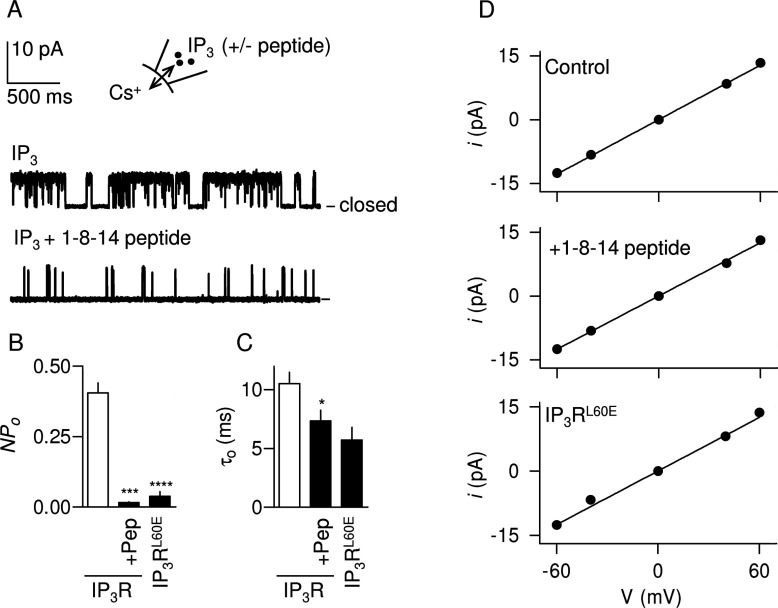
In patch-clamp recordings from the nuclear envelope of DT40 cells expressing rat IP3R1, a maximally effective concentration of IP3 stimulated IP3R activity and this was massively attenuated by the 1-8-14 peptide (3 mM) (Figures 3A and 3B). Our results are consistent with the peptide causing a 50% decrease in the mean channel open time (τo) (Figure 3C). However, the overall channel activity (NPo) was so low under these conditions that we cannot reliably estimate the number of active IP3Rs (N) within each patch. We cannot therefore entirely eliminate the possibility that each patch fortuitously included several IP3Rs and that their clustering caused τo to fall from ~10 ms to ~5 ms as we reported previously [40]. An effect on τo would be unusual because most regulators of IP3Rs affect the duration of closed states (τc) [6,40]. The effect of the peptide on τo is not, however, sufficient to account for the ~10-fold decrease in NPo (Figure 3A), suggesting that the 1-8-14 peptide must also affect the rate of channel opening (i.e. τc). Because it was impossible to determine the number of active IP3Rs in the presence of 1-8-14 peptide (see above), we could not reliably determine τc. The single-channel conductance (γCs) was unaffected by 1-8-14 peptide: it was 214±6 pS (n=3) and 209±6 pS (n=3) for control and peptide-treated IP3Rs respectively (Figure 3D).
Figure 3. Inhibition of IP3R gating by 1-8-14 peptide.
(A) Typical recordings from excised nuclear patches stimulated with IP3 (10 μM) with and without 1-8-14 peptide (3 mM) in the pipette solution. The holding potential was +40 mV. The closed state is shown. (B and C) NPo (B) and τo (C) for IP3R stimulated with IP3 alone or with 1-8-14 peptide (3 mM, +Pep). Results for IP3RL60E are also shown. *P<0.05, ***P<0.001 and ****P<0.0001 relative to native IP3R without peptide. (D) Single-channel current (i)–voltage (V) relationships for the three stimulation conditions. Results in (B)–(D) are means±S.E.M. (n≥3).
These results establish that a peptide derived from an endogenous 1-8-14 motif within the SD of the IP3R is similar to MLCK peptide in causing substantial and reversible inhibition of IP3Rs that is independent of CaM. This conclusion is consistent with our suggestion that MLCK peptide inhibits IP3Rs by mimicking an endogenous 1-8-14 motif, and so perhaps ‘unzipping’ an interdomain interaction [43] that is essential for activation of IP3Rs.
1-8-14 peptide uncouples IP3 binding from activation of IP3Rs
Removal of the SD increases the affinity of both full-length IP3Rs and the NT for IP3 [33]. We [33] have suggested that this reflects the use of binding energy to drive conformational rearrangement of SD-IBC interfaces during the initial steps of IP3R activation [5,44].
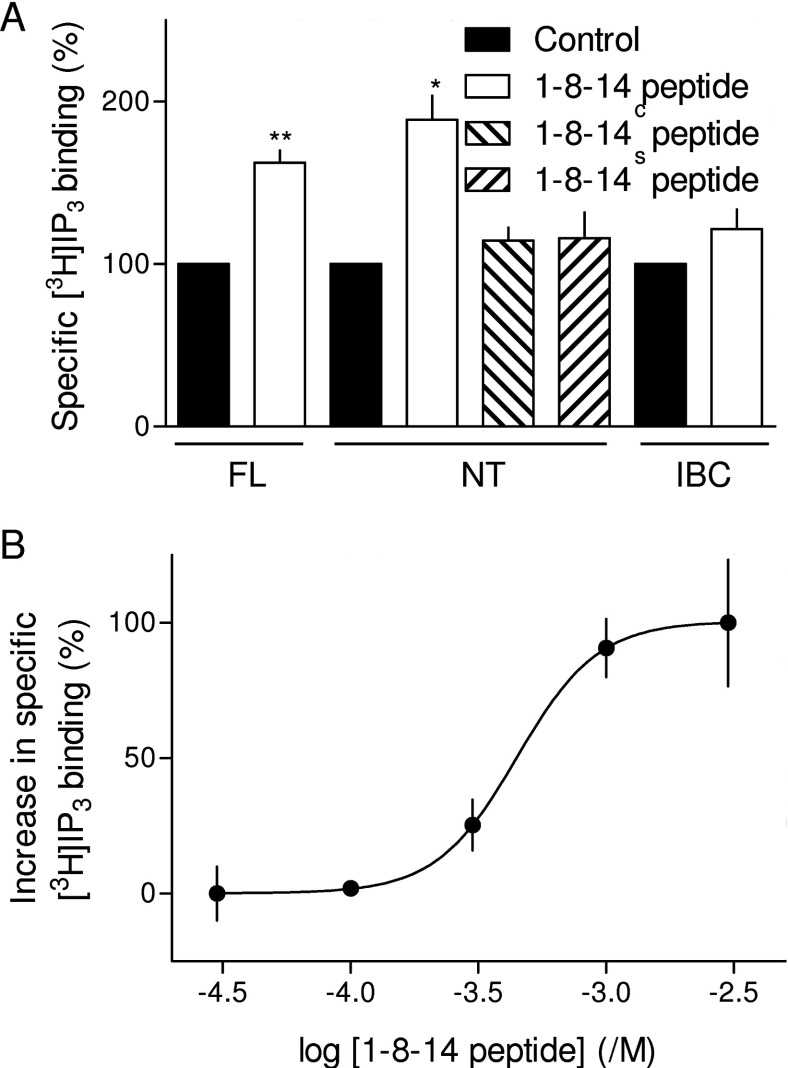
1-8-14 peptide (3 mM) increased specific binding of [3H]IP3 to full-length IP3R1. Similar results were obtained with the NT, but IP3 binding to the IBC was unaffected (Figure 4A). The latter demonstrates that 1-8-14 peptide does not interact directly with either the IP3-binding site or with IP3. Neither the mutated (1-8-14C) nor scrambled (1-8-14S) peptide had any effect on IP3 binding to the NT (Figure 4A). These results with IP3R fragments expressed in E. coli, which lack CaM, also further support our conclusion that the effects of 1-8-14 peptide are entirely independent of CaM.
Figure 4. 1-8-14 peptide directly stimulates IP3 binding to the NT of IP3R.
(A) Specific equilibrium binding of [3H]IP3 (1.5 nM) to membranes from rat cerebellum (full-length IP3R, FL) or to isolated NT or IBC, alone or in the presence of 3 mM of the indicated peptide. *P<0.05 and **P<0.01 relative to control; comparisons were performed on the raw data. (B) Concentration-dependent effects of 1-8-14 peptide on specific [3H]IP3 binding to NT in CLM with 220 nM free Ca2+ concentration, plotted as the increase in specific [3H]IP3 binding as a percentage of that evoked by the maximal concentration of peptide. Results are means±S.E.M. (n≥3).
Comparison of the effects of 1-8-14 peptide on stimulating [3H]IP3 binding to the NT (EC50, 615 μM; pEC50, 3.21±0.19) (Figure 4B) with its inhibitory effect on IP3-evoked Ca2+ release (IC50, 767 μM; pIC50, 3.1±0.25) (Figure 2C) demonstrates that each is similarly sensitive to the peptide. These results are consistent with our hypothesis that the 1-8-14 peptide disrupts an interaction between the SD and IBC that is essential for IP3R activation. The peptide thereby inhibits IP3-evoked Ca2+ release (Figure 2) and IP3R activity (Figure 3) and, by uncoupling IP3 binding from subsequent conformational changes, it stimulates IP3 binding (Figure 4). Subsequent experiments used mutagenesis of residues within the endogenous 1-8-14 motif to test this hypothesis further.
Mutations within the endogenous 1-8-14 sequence increase IP3-binding affinity
If, as we suggest, the 1-8-14 peptide disrupts an essential interaction between the endogenous 1-8-14 sequence and another domain within the NT, we might expect mutation of appropriate residues in the SD to both disrupt IP3R activation and increase IP3-binding affinity. We tested the latter prediction by examining IP3 binding to the NT in which each of the critical (1, 8 and 14) hydrophobic/aromatic residues that are important for Ca2+–CaM binding to 1-8-14 motifs [45] was replaced with a charged hydrophilic residue (glutamate). The same hydrophobic residues are essential for MLCK [35] and 1-8-14 (Figure 2C) peptides to disrupt IP3R activation.
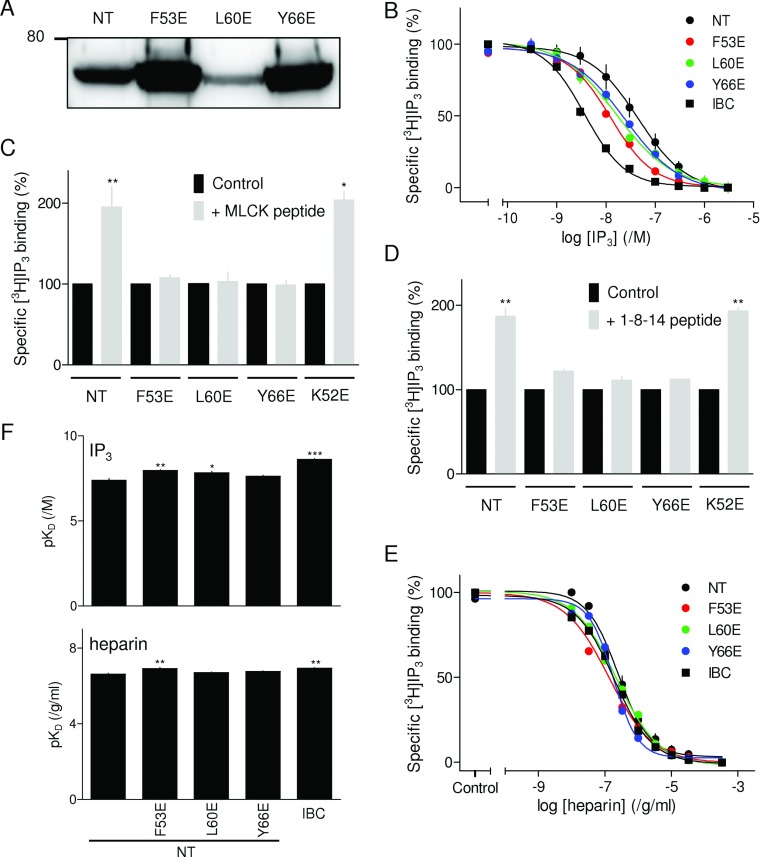
NTs of IP3R1 with point mutations in positions equivalent to the 1- (F53E), 8- (L60E) or 14-position (Y66E) of the endogenous 1-8-14 motif (Figure 1A) were expressed in E. coli. Expression levels of the NT and its mutants were not identical (Figure 5A), but they were each sufficient to allow the affinity for IP3 and the effects of peptides to be determined after cleavage of the His6 tag, but without further purification [33]. As expected, IP3 bound to the IBC with greater affinity (17-fold) than to the NT (Figure 5B) [33,46,47], consistent with our suggestion that, in the absence of the SD, less binding energy is diverted into conformational changes [33]. Mutation of critical residues within the endogenous 1-8-14 motif significantly increased the affinity of the NT for IP3 (Figure 5B and Table 1), although none was as effective as complete removal of the SD. This is consistent with our observation that neither the 1-8-14 (Figure 2) nor MLCK [35] peptide entirely inhibits IP3-evoked Ca2+ release, whereas removal of the SD totally uncouples IP3 binding from IP3R activation [48]. Although maximally effective concentrations of MLCK (100 μM) or 1-8-14 (3 mM) peptides similarly increased IP3 binding to the NT, neither peptide had any effect on [3H]IP3 binding to the NT with mutations in any of the critical 1-8-14 residues (Figures 5C and 5D). Mutation of a residue immediately preceding the critical 1-position of the 1-8-14 motif (K52E), which did not increase the affinity of IP3 for the NT (Supplementary Figure S2A at http://www.BiochemJ.org/bj/449/bj4490039add.htm), had no effect on the responses to MLCK or 1-8-14 peptides (Figures 5C and 5D) and neither did it affect IP3-evoked Ca2+ release [33] (Supplementary Figure S2B). These results establish that mutation of critical residues within the endogenous 1-8-14 motif selectively increases IP3-binding affinity and these effects are non-additive with those of either MLCK or 1-8-14 peptide.
Figure 5. Mutations within the 1-8-14 motif mimic the effect of 1-8-14 peptide on IP3 binding.
(A) Western blot (typical of three independent experiments) with an anti-His6 antibody of lysates (5 μg of protein/lane) from bacteria expressing NT with the indicated mutations. The 80 kDa molecular-mass marker is shown. (B) Concentration-dependent effect of IP3 on specific [3H]IP3 binding to the IBC, NT and mutated NT. (C and D) Effects of MLCK peptide (C, 100 μM) and 1-8-14 peptide (D, 3 mM) on specific binding of [3H]IP3 (1.5 nM) to the NT and the indicated mutants (each expressed as a percentage of the control). (E) Specific binding of [3H]IP3 (1.5 nM) to the IBC, NT and mutated NT in the presence of the indicated concentrations of heparin. (F) Summary results from experiments similar to those in (E) showing the Kd for IP3 and heparin binding to the IBC, NT and mutated NT. Results in (B)–(F) are means±S.E.M. (n≥3). *P<0.05, **P<0.01 and ***P<0.001 relative to control; comparisons were performed on the raw data.
Table 1. Binding of IP3 and heparin to N-terminal fragments of IP3R1.
Equilibrium competition binding using [3H]IP3 was used to measure the pKd of IP3 and heparin for the N-terminal fragments of IP3R1. Affinities for ligands are also shown expressed as fold increase relative to wild-type NT (i.e. KdNT/Kdmutant). Results are means±S.E.M. (n≥3). *P<0.05, **P<0.01 and ***P<0.001 relative to NT.
| Fragment | pKd, /M IP3 (Kd, nM) | Affinity relative to NT | pKd, /g/ml heparin (Kd, ng/ml) | Affinity relative to NT |
|---|---|---|---|---|
| NT | 7.40±0.11 (40.0) | 1 | 6.62±0.06 (239) | 1 |
| F53E | 7.97±0.05** (10.8) | 4 | 6.92±0.06** (120) | 2 |
| L60E | 7.84±0.08* (14.5) | 3 | 6.70±0.04 (200) | 1.2 |
| Y66E | 7.64±0.04 (22.8) | 2 | 6.77±0.03 (171) | 1.4 |
| IBC | 8.62±0.05*** (2.4) | 17 | 6.93±0.04** (117) | 2.0 |
Mutations within the 1-8-14 motif selectively increase agonist affinity
Our hypothesis is that the apparent affinity of agonists (such as IP3) for native IP3Rs is reduced because some of their binding energy is diverted into the conformational changes that activate the IP3R [33]. Antagonists, because they need not evoke the rearrangement of the IBC and SD that initiates IP3R activation, may be less affected by disruption of these interactions. We therefore examined the effects of the SD and of point mutations within the endogenous 1-8-14 sequence on binding to the NT of heparin, a competitive antagonist of IP3 [49]. The results demonstrate that, whereas removal of the SD increased the affinity of the NT for IP3 17-fold, it caused only a 2-fold increase in the affinity for heparin. Point mutations within the endogenous 1-8-14 motif also caused larger increases in the affinity for IP3 than for heparin (Figures 5E and 5F, and Table 1). These results are important because they demonstrate that the effects of the SD and of mutations within the 1-8-14 sequence on ligand binding are specific for an agonist of the IP3R. They thereby demonstrate the importance of the 1-8-14 motif in specifically mediating activation of IP3Rs.
Mutations within the endogenous 1-8-14 motif uncouple IP3 binding from gating of IP3Rs
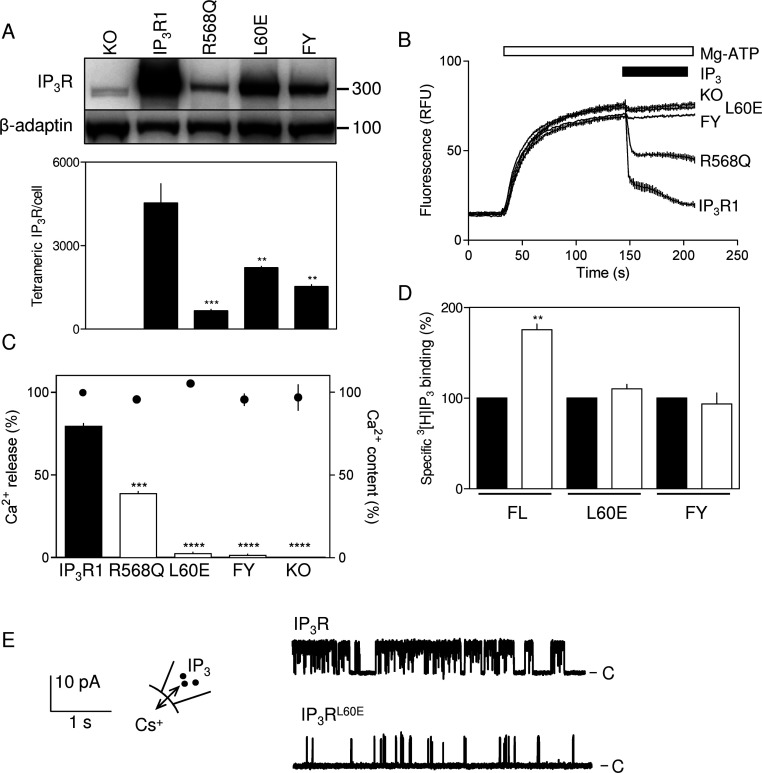
It proved difficult to establish stable DT40 cell lines expressing rat IP3R1 in which critical residues within the 1-8-14 motif were mutated, but we succeeded with two mutants (Figure 6A). The first (IP3RL60E) is mutated at the 8-position of the 1-8-14 motif and the second has mutations at both the 1- (F53E) and 14-positions (Y66E) (IP3RFY). As expected, a maximally effective concentration of IP3 (10 μM) failed to stimulate Ca2+ release from permeabilized DT40 cells lacking IP3R (DT40-KO cells) [37,40], but it caused release of 81±1% of the Ca2+ stores of DT40-IP3R1 cells (Figures 6B and 6C). In the cell lines expressing IP3R with a mutated 1-8-14 motif, there was barely detectable Ca2+ release that was not significantly different from that observed in DT40-KO cells (Figures 6B and 6C). ATP-dependent Ca2+ uptake into the ER was similar for each cell line (Figure 6C). We were concerned that the lower level of expression of mutant IP3R relative to wild-type (~30–50%, Figure 6A) might have contributed to the lack of detectable IP3-evoked Ca2+ release. However, in another stable DT40 cell line where the IP3-binding site was mutated (R568Q), causing a ~10-fold decrease in IP3 affinity [50], IP3R expression (~15% of wild-type) was less than half that of the cell lines with mutations in the 1-8-14 motif (Figure 6A). Nevertheless, IP3 caused a readily detectable release of Ca2+ from the intracellular stores of DT40-IP3RR568Q cells (49±2% of that detected in DT40-IP3R1 cells) (Figures 6B and 6C). We conclude that the lack of detectable Ca2+ release in cells expressing IP3R with a mutant 1-8-14 motif is not attributable to reduced IP3R expression. Neither is it likely that the lack of response to IP3 from mutant IP3R reflects a more global disruption of IP3R structure because each of the full-length mutant IP3Rs bound IP3, although, as predicted, addition of MLCK peptide increased IP3 binding to only the wild-type IP3R (Figure 6D). Furthermore, DT40 cells expressing IP3R1 with a mutation in an adjacent residue (DT40-IP3R1K52E) responded normally to IP3 [33] (Supplementary Figure S2B). These results are consistent with the suggestion that mutations within the endogenous 1-8-14 motif mimic addition of exogenous MLCK peptide by uncoupling IP3 binding from the conformational changes that lead to opening of the IP3R pore. Single-channel analyses provide further support for this conclusion.
Figure 6. The endogenous 1-8-14 motif is essential for activation of IP3R.
(A) Expression of IP3R1 in DT40 cells stably expressing each of the indicated mutants. Each lane was loaded with 4×103 cells and probed with antisera to IP3R1 (upper panel) or β-adaptin (lower panel). The R568Q mutant (which reduces the affinity of the IP3R for IP3) [50] is shown because it provides a control for functional assays of cells expressing IP3R at low density. Molecular-mass markers are shown on the right. The Western blot is typical of three independent experiments. The lower panel shows summary results (means±S.E.M., n=3), where IP3R expression was calculated from blots that included DT40-IP3R1 membranes in which levels of expression were established by equilibrium competition [3H]IP3 binding. (B) Typical responses to IP3 (10 μM) from DT40 cells lacking IP3R (KO) or expressing wild-type IP3R1 or IP3R with the indicated mutations (see the text for details). (C) Summary results show the Ca2+ content of the loaded stores (●) and the Ca2+ released by IP3 (histograms) for each of the indicated cell lines. (D) Specific [3H]IP3 binding (1.5 nM) to full-length IP3R (FL) with the indicated mutations (L60E or FY, see the text for details) in permeabilized DT40 cells alone or in the presence of 100 μM MLCK peptide. Results in (C) and (D) are means±S.E.M. (n≥3). (E) Typical records from active excised nuclear patches of DT40 cells expressing IP3R1 or IP3R1L60E stimulated with IP3 (10 μM). The holding potential was +40 mV. C denotes the closed state. Summary data are provided in Figures 3(B)–3(D). **P<0.01, ***P<0.001 and ****P<0.0001 relative to IP3R1 (A and C) or control (D).
Yamazaki et al. [51] reported recently the functional effects of mutations within IP3R including some within the 1-8-14 motif (F53D and Y66A). We note, however, that some of their mutations, e.g. Y167A, which is clearly implicated in IP3R activation, abolished IP3-evoked Ca2+ release from microsomes without affecting Ca2+ signals evoked by activation of the BCR (B-cell receptor) in intact cells. This unexplained disparity casts some doubt over whether in these assays responses from intact cells faithfully report the activity of IP3R. In DT40 cells expressing an IP3R with five mutations that included Y66A (the 14-position of the 1-8-14 motif), activation of the BCR evoked a Ca2+ signal, suggesting that the mutant IP3R was functional [51]. However, in this IP3R, the mutant had one hydrophobic residue replaced by another and this might not radically affect the behaviour of the 1-8-14 motif. In preliminary analyses of cells expressing IP3Rs in which the first position of the 1-8-14 motif was mutated (F53D), Ca2+ signals were also observed after activation of the BCR [51]. This may reflect a limitation of the BCR-based assay (see above) or it may provide evidence for a lesser role of the 1-position in the 1-8-14 motif. We have not succeeded in establishing a DT40 cell line expressing IP3Rs with only this mutation, although our results do clearly show that IP3Rs with mutations in both the 1- and 14-positions (IP3RFY) are barely responsive to IP3 (Figure 6).
Mutation of the endogenous 1-8-14 motif attenuates IP3R gating without affecting single-channel conductance
In keeping with the reduced expression of IP3RL60E in DT40 cells (Figure 6A), the frequency with which functional IP3Rs were detected in excised nuclear patches was much lower for nuclei from paired experiments with DT40-IP3R1L60E cells (three of 48 patches) than from DT40-IP3R1 cells (five of 13 patches). In parallel analyses, functional IP3Rs were never detected in DT40-KO cells (none of 30 patches). The single-channel conductances (γCs) of the mutant IP3RL60E (209±8 pS) and normal IP3R (214±6 pS) were indistinguishable (Figure 3D), but NPo was massively decreased in the mutant (Figures 3B and 6E). Our interpretation of the latter is, as we described in our analyses of the 1-8-14 peptide, limited by our inability, when NPo is so low for IP3R1L60E, to estimate reliably the number of active IP3R within a patch. Nevertheless, it is clear that the major effect on single-channel behaviour of mutating the endogenous 1-8-14 motif of IP3R1 (Figures 3B–3D and 6E) and of adding 1-8-14 peptide to normal IP3R1 (Figure 3) is similar: both decrease NPo without affecting γCs. These results establish that mutations in the endogenous 1-8-14 motif or addition of 1-8-14 peptide uncouple ligand binding from channel gating without compromising the behaviour of the pore.
Conclusions: interactions between endogenous 1-8-14 and CaM-like motifs mediate activation of IP3Rs
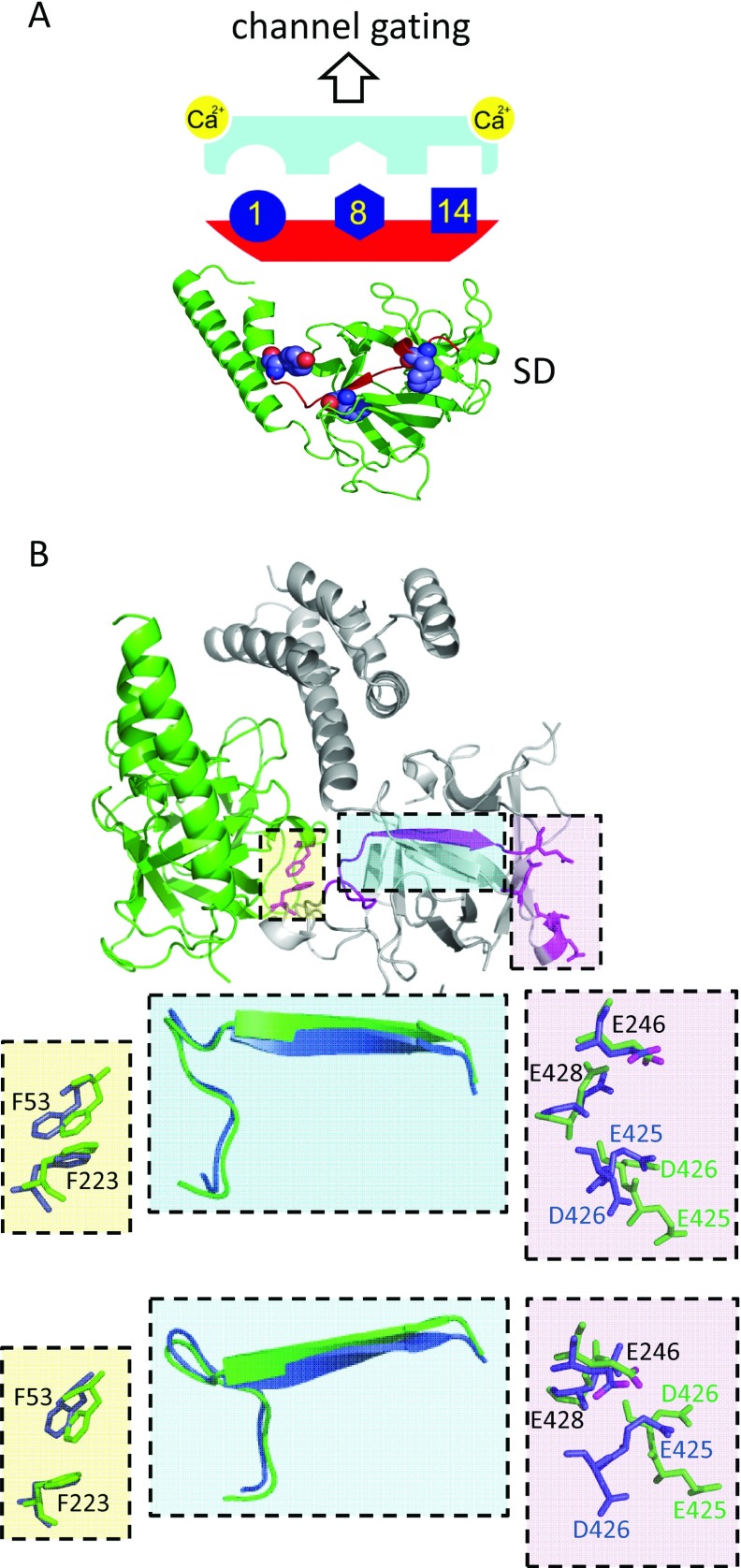
CaM [22] or related EF-hand-containing proteins [14,25], peptides that comprise 1-8-14 CaM-binding motifs [35,36] (Figures 2–4) or disruption of a conserved endogenous 1-8-14-like motif within the SD of IP3Rs inhibit IP3-evoked Ca2+ release (Figures 5 and 6) by massively reducing NPo of IP3R (Figures 3 and 6E). We conclude that an endogenous 1-8-14 motif within the SD (Figure 1) is essential for IP3R activation. Where it has been examined, the inhibitory proteins or peptides are more potent when Ca2+ is bound to the IP3R [26,35] (Figure 2F). We therefore speculate that the endogenous 1-8-14 motif may interact with an unidentified domain that includes an EF-hand-like structure and that these interactions might be related to Ca2+ regulation of IP3R (Figure 7). We suggest that competing peptides (CaM-like or 1-8-14 motifs) or mutagenesis of the endogenous 1-8-14 motif inhibit IP3Rs by disrupting this essential interaction in a manner similar to the ‘unzipping’ of interdomain interactions in RyRs [32,43,52]. The scheme is appealing because IP3 regulates binding of Ca2+ to IP3Rs and thereby leads to channel gating [34,53]. The identity of this Ca2+-binding site is unknown. It is, however, clear that Ca2+ regulates IP3 binding to the NT only when the SD is present [42], suggesting that a Ca2+-binding site within the NT may be regulated by interactions between the SD and IBC. One possibility is that an endogenous EF-hand-like structure might provide the Ca2+-binding site and that its interaction with the 1-8-14 motif links IP3 and Ca2+ binding (Figure 7A). Bioinformatic analyses had suggested the presence of two possible EF-hand-like structures within the IBC [9,54], but neither is evident in high-resolution structures of the IBC [55] and NT [5,56]. Neither have we succeeded in identifying a complementary partner of the 1-8-14 motif. Another possibility is suggested by comparison of the structures of the NT with and without IP3 bound [5,56], which reveal that Phe53 (the first hydrophobic residue of the 1-8-14 motif) and Phe223 are closely apposed (~3.9 Å; 1 Å=0.1 nm), but they move apart (~5.3 Å) when IP3 binds (Figure 7B). A β-sheet links Phe223 to Glu246, and the movement of Phe223 is associated with a repositioning of an acidic residue in the β-domain of the IBC (Glu246). This brings Glu246 closer to three other acidic residues (Glu425, Asp426 and Glu428). The rearrangement is interesting because these four residues have been proposed to form a Ca2+-binding site (Ca-I) [55]. Furthermore, a peptide (residues 378–450) that includes most of these residues binds Ca2+, and the binding is abolished by mutation of the acidic residues [42]. A second possibility is therefore that IP3-evoked movement of the critical 1-8-14 motif contributes to formation of an effective Ca2+-binding site within the IBC by bringing a fourth acidic residue into appropriate association with three others.
Figure 7. Activation of IP3Rs requires an endogenous 1-8-14 motif.
IP3 binding to the IBC initiates conformational changes that pass via the SD and lead, via regulation of Ca2+ binding to the IP3R, to opening of the pore [33]. (A) An endogenous 1-8-14 motif within the SD is essential for IP3R activation. We speculate (upper panel) that interaction of this CaM-binding motif (red, conserved hydrophobic residues in dark blue) with an endogenous, but presently unknown, CaM-like structure (pale blue) within the NT may link IP3 binding to Ca2+ binding. (B) Another possibility is that IP3 binding rearranges the 1-8-14 motif and so repositions a critical acidic residue (Glu246) that may then contribute to a Ca2+-binding site (Ca-1) [55]. The NT without IP3 bound (PDB code 3UJ0) [5] is shown with the IBC in grey and the SD in green to highlight Phe53 (within the 1-8-14 motif) and Phe223 to which it is closely apposed (yellow box), residues proposed to form the Ca-1 site (pink box) and the β-sheet that links Phe223 to Glu246 (cyan box). The expanded views (each rotated to show key movements) show the critical residues and the linking β-sheet before (green) and after IP3 binding (blue, PDB code 3UJ4). The carboxy oxygen atoms in Glu246 are shown in magenta. We speculate that separation of Phe53 and Phe223 when IP3 binds is associated with twisting of the linking β-sheet and movement of Glu246 towards three other acidic residues (Glu425, Asp426 and Glu428) and that they may then together form an effective Ca2+-binding site.
We conclude that a conserved 1-8-14 motif within the SD is essential for IP3R activation and speculate that its interaction with either an endogenous CaM-like motif or acidic residues within the IBC may link IP3 and Ca2+ binding. Inhibition of IP3R by CaM and related proteins probably results from disruption of this essential interaction.
Online data
AUTHOR CONTRIBUTION
Yi Sun and Ana Rossi performed the Ca2+-release and IP3-binding analyses. Taufiq Rahman performed the single-channel analyses. Colin Taylor directed the study, and with input from all authors, wrote the paper.
FUNDING
Supported by the Wellcome Trust [grant number 085295], Biotechnology and Biological Sciences Research Council [grant number BB/H009736/1] and a studentship from the Engineering and Physical Sciences Research Council (to Y.S.). A.R. is a fellow of Queens’ College, Cambridge. T.R. is a Drapers Research Fellow of Pembroke College, Cambridge.
References
- 1.Berridge M. J., Lipp P., Bootman M. D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Tadross M. R., Dick I. E., Yue D. T. Mechanism of local and global Ca2+ sensing by calmodulin in complex with a Ca2+ channel. Cell. 2008;133:1228–1240. doi: 10.1016/j.cell.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchant J. S., Parker I. Role of elementary Ca2+ puffs in generating repetitive Ca2+ oscillations. EMBO J. 2001;20:65–76. doi: 10.1093/emboj/20.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton S. L., Serysheva I. I. Ryanodine receptor structure: progress and challenges. J. Biol. Chem. 2009;284:4047–4051. doi: 10.1074/jbc.R800054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seo M.-D., Velamakanni S., Ishiyama N., Stathopulos P. B., Rossi A. M., Khan S. A., Dale P., Li C., Ames J. B., Ikura M., Taylor C. W. Structural and functional conservation of key domains in InsP3 and ryanodine receptors. Nature. 2012;483:108–112. doi: 10.1038/nature10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foskett J. K., White C., Cheung K. H., Mak D. O. Inositol trisphosphate receptor Ca2+ release channels. Physiol. Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zalk R., Lehnart S. E., Marks A. R. Modulation of the ryanodine receptor and intracellular calcium. Annu. Rev. Biochem. 2007;76:367–385. doi: 10.1146/annurev.biochem.76.053105.094237. [DOI] [PubMed] [Google Scholar]
- 8.Chen S. R. W., MacLennan D. H. Identification of calmodulin-, Ca2+- and ruthenium red-binding domains in the Ca2+ release channel (ryanodine receptor) of rabbit skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 1994;269:22698–22704. [PubMed] [Google Scholar]
- 9.Sienaert I., Missiaen L., De Smedt H., Parys J. B., Sipma H., Casteels R. Molecular and functional evidence for multiple Ca2+-binding domains on the type 1 inositol 1,4,5-trisphosphate receptor J. Biol. Chem. 1997;272:25899–25906. doi: 10.1074/jbc.272.41.25899. [DOI] [PubMed] [Google Scholar]
- 10.Fessenden J. D., Feng W., Pessah I. N., Allen P. D. Mutational analysis of putative calcium binding motifs within the skeletal ryanodine receptor isoform, RyR1. J. Biol. Chem. 2004;279:53028–53035. doi: 10.1074/jbc.M411136200. [DOI] [PubMed] [Google Scholar]
- 11.Miyakawa T., Mizushima A., Hirose K., Yamazawa T., Bezprozvanny I., Kurosaki T., Iino M. Ca2+-sensor region of IP3 receptor controls intracellular Ca2+ signaling. EMBO J. 2001;20:1674–1680. doi: 10.1093/emboj/20.7.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin D., Means A. R. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 13.Wright N. T., Prosser B. L., Varney K. M., Zimmer D. B., Schneider M. F., Weber D. J. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J. Biol. Chem. 2008;283:26676–26683. doi: 10.1074/jbc.M804432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadif Kasri N., Holmes A. M., Bultynck G., Parys J. B., Bootman M. D., Rietdorf K., Missiaen L., McDonald F., De Smedt H., Conway S. J., et al. Regulation of InsP3 receptor activity by neuronal Ca2+-binding proteins. EMBO J. 2004;23:312–321. doi: 10.1038/sj.emboj.7600037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michikawa T., Hirota J., Kawano S., Hiraoka M., Yamada M., Furuichi T., Mikoshiba K. Calmodulin mediates calcium-dependent inactivation of the cerebellar type 1 inositol 1,4,5-trisphosphate receptor. Neuron. 1999;23:799–808. doi: 10.1016/s0896-6273(01)80037-4. [DOI] [PubMed] [Google Scholar]
- 16.Nosyreva E., Miyakawa T., Wang Z., Glouchankova L., Iino M., Bezprozvanny I. The high-affinity calcium–calmodulin-binding site does not play a role in the modulation of type 1 inositol 1,4,5-trisphosphate receptor function by calcium and calmodulin. Biochem. J. 2002;365:659–667. doi: 10.1042/BJ20011789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X., Joseph S. K. Effect of mutation of a calmodulin-binding sites on Ca2+ regulation of inositol trisphosphate receptors. Biochem. J. 2001;360:395–400. doi: 10.1042/0264-6021:3600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor C. W., Laude A. J. IP3 receptors and their regulation by calmodulin and cytosolic Ca2+ Cell Calcium. 2002;32:321–334. doi: 10.1016/s0143416002001859. [DOI] [PubMed] [Google Scholar]
- 19.Rodney G. G., Moore C. P., Williams B. Y., Zhang J.-Z., Krol J., Pedersen S. E., Hamilton S. L. Calcium binding to calmodulin leads to an N-terminal shift in its binding site on the ryanodine receptor. J. Biol. Chem. 2001;276:2069–2074. doi: 10.1074/jbc.M008891200. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi N., Takahashi N., Xu L., Smithies O., Meissner G. Early cardiac hypertrophy in mice with impaired calmodulin regulation of cardiac muscle Ca2+ release channel. J. Clin. Invest. 2007;117:1344–1353. doi: 10.1172/JCI29515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Missiaen L., Parys J. B., Weidema A. F., Sipma H., Vanlingen S., De Smet P., Callewaert G., De Smedt H. The bell-shaped Ca2+-dependence of the inositol 1,4,5-trisphosphate induced Ca2+ release is modulated by Ca2+/calmodulin. J. Biol. Chem. 1999;274:13748–13751. doi: 10.1074/jbc.274.20.13748. [DOI] [PubMed] [Google Scholar]
- 22.Adkins C. E., Morris S. A., De Smedt H., Török K., Taylor C. W. Ca2+-calmodulin inhibits Ca2+ release mediated by type-1, -2 and -3 inositol trisphosphate receptors. Biochem. J. 2000;345:357–363. [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C., Widjaja J., Joseph S. K. The interaction of calmodulin with alternatively spliced isoforms of the type-I inositol trisphosphate receptor. J. Biol. Chem. 2000;275:2305–2311. doi: 10.1074/jbc.275.4.2305. [DOI] [PubMed] [Google Scholar]
- 24.Yamada M., Miyawaki A., Saito K., Yamamoto-Hino M., Ryo Y., Furuichi T., Mikoshiba K. The calmodulin-binding domain in the mouse type 1 inositol 1,4,5-trisphosphate receptor. Biochem. J. 1995;308:83–88. doi: 10.1042/bj3080083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C., Chan J., Haeseleer F., Mikoshiba K., Palczewski K., Ikura M., Ames J. B. Structural insights into Ca2+-dependent regulation of inositol 1,4,5-trisphosphate receptors by CaBP1. J. Biol. Chem. 2009;284:2472–2481. doi: 10.1074/jbc.M806513200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadif Kasri N., Bultynck G., Smyth J., Szlufcik K., Parys J., Callewaert G., Missiaen L., Fissore R. A., Mikoshiba K., De Smedt H. The N-terminal Ca2+-independent calmodulin-binding site on the inositol 1,4,5-trisphosphate receptor is responsible for calmodulin inhibition, even though this inhibition requires Ca2+ Mol. Pharmacol. 2004;66:276–284. doi: 10.1124/mol.66.2.276. [DOI] [PubMed] [Google Scholar]
- 27.Rossi A., Taylor C. W. Ca2+ regulation of inositol 1,4,5-trisphosphate receptors: can Ca2+ function without calmodulin? Mol. Pharmacol. 2004;66:199–203. doi: 10.1124/mol.104.002592. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi N., Xu L., Evans K. E., Pasek D. A., Meissner G. Different regions in skeletal and cardiac muscle ryanodine receptors are involved in transducing the functional effects of calmodulin. J. Biol. Chem. 2004;279:36433–36439. doi: 10.1074/jbc.M405834200. [DOI] [PubMed] [Google Scholar]
- 29.Rodney G. G., Wilson G. M., Schneider M. F. A calmodulin binding domain of RyR increases activation of spontaneous Ca2+ sparks in frog skeletal muscle. J. Biol. Chem. 2005;280:11713–11722. doi: 10.1074/jbc.M408189200. [DOI] [PubMed] [Google Scholar]
- 30.Sencer S., Papineni R. V., Halling D. B., Pate P., Krol J., Zhang J. Z., Hamilton S. L. Coupling of RYR1 and L-type calcium channels via calmodulin binding domains. J. Biol. Chem. 2001;276:38237–38241. doi: 10.1074/jbc.C100416200. [DOI] [PubMed] [Google Scholar]
- 31.Xiong L., Zhang J. Z., He R., Hamilton S. L. A Ca2+-binding domain in RyR1 that interacts with the calmodulin binding site and modulates channel activity. Biophys. J. 2006;90:173–182. doi: 10.1529/biophysj.105.066092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu X., Ghanta J., Walker J. W., Allen P. D., Valdivia H. H. The calmodulin binding region of the skeletal ryanodine receptor acts as a self-modulatory domain. Cell Calcium. 2004;35:165–177. doi: 10.1016/j.ceca.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Rossi A. M., Riley A. M., Tovey S. C., Rahman T., Dellis O., Taylor E. J. A., Veresov V. G., Potter B. V. L., Taylor C. W. Synthetic partial agonists reveal key steps in IP3 receptor activation. Nat. Chem. Biol. 2009;5:631–639. doi: 10.1038/nchembio.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchant J. S., Taylor C. W. Cooperative activation of IP3 receptors by sequential binding of IP3 and Ca2+ safeguards against spontaneous activity. Curr. Biol. 1997;7:510–518. doi: 10.1016/s0960-9822(06)00222-3. [DOI] [PubMed] [Google Scholar]
- 35.Sun Y., Taylor C. W. A calmodulin antagonist reveals a calmodulin-independent interdomain interaction essential for activation of inositol 1,4,5-trisphosphate receptors. Biochem. J. 2008;416:243–253. doi: 10.1042/BJ20080861. [DOI] [PubMed] [Google Scholar]
- 36.Kasri N. N., Török K., Galione A., Garnham C., Callewaert G., Missiaen L., Parys J. B., De Smedt H. Endogenously bound calmodulin is essential for the function of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 2006;281:8332–8338. doi: 10.1074/jbc.M510971200. [DOI] [PubMed] [Google Scholar]
- 37.Sugawara H., Kurosaki M., Takata M., Kurosaki T. Genetic evidence for involvement of type 1, type 2 and type 3 inositol 1,4,5-trisphosphate receptors in signal transduction through the B-cell antigen receptor. EMBO J. 1997;16:3078–3088. doi: 10.1093/emboj/16.11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cardy T. J. A., Traynor D., Taylor C. W. Differential regulation of types 1 and 3 inositol trisphosphate receptors by cytosolic Ca2+ Biochem. J. 1997;328:785–793. doi: 10.1042/bj3280785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tovey S. C., Sun Y., Taylor C. W. Rapid functional assays of intracellular Ca2+ channels. Nat. Protoc. 2006;1:259–263. doi: 10.1038/nprot.2006.40. [DOI] [PubMed] [Google Scholar]
- 40.Rahman T. U., Skupin A., Falcke M., Taylor C. W. Clustering of IP3 receptors by IP3 retunes their regulation by IP3 and Ca2+ Nature. 2009;458:655–659. doi: 10.1038/nature07763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bosanac I., Yamazaki H., Matsu-ura T., Michikawa M., Mikoshiba K., Ikura M. Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor. Mol. Cell. 2005;17:193–203. doi: 10.1016/j.molcel.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 42.Sienaert I., Kasri N. N., Vanlingen S., Parys J., Callewaert G., Missiaen L., De Smedt H. Localization and function of a calmodulin/apocalmodulin binding domain in the N-terminal part of the type 1 inositol 1,4,5-trisphosphate receptor. Biochem. J. 2002;365:269–277. doi: 10.1042/BJ20020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikemoto N., Yamamoto T. Regulation of calcium release by interdomain interaction within ryanodine receptors. Front. Biosci. 2002;7:671–683. doi: 10.2741/A803. [DOI] [PubMed] [Google Scholar]
- 44.Chan J., Whitten A. E., Jeffries C. M., Bosanac I., Mal T. K., Ito J., Porumb H., Michikawa T., Mikoshiba K., Trewhella J., Ikura M. Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor. J. Mol. Biol. 2007;373:1269–1280. doi: 10.1016/j.jmb.2007.08.057. [DOI] [PubMed] [Google Scholar]
- 45.Rhoads A. R., Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 46.Yoshikawa F., Morita M., Monkawa T., Michikawa T., Furuichi T., Mikoshiba K. Mutational analysis of the ligand binding site of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 1996;271:18277–18284. doi: 10.1074/jbc.271.30.18277. [DOI] [PubMed] [Google Scholar]
- 47.Iwai M., Michikawa T., Bosanac I., Ikura M., Mikoshiba K. Molecular basis of the isoform-specific ligand-binding affinity of inositol 1,4,5-trisphosphate receptors. J. Biol. Chem. 2007;282:12755–12764. doi: 10.1074/jbc.M609833200. [DOI] [PubMed] [Google Scholar]
- 48.Uchida K., Miyauchi H., Furuichi T., Michikawa T., Mikoshiba K. Critical regions for activation gating of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 2003;278:16551–16560. doi: 10.1074/jbc.M300646200. [DOI] [PubMed] [Google Scholar]
- 49.Ghosh T. K., Eis P. S., Mullaney J. M., Ebert C. L., Gill D. L. Competitive, reversible, and potent antagonism of inositol 1,4,5-trisphosphate-activated calcium release by heparin. J. Biol. Chem. 1988;263:11075–11079. [PubMed] [Google Scholar]
- 50.Dellis O., Rossi A. M., Dedos S. G., Taylor C. W. Counting functional IP3 receptors into the plasma membrane. J. Biol. Chem. 2008;283:751–755. doi: 10.1074/jbc.M706960200. [DOI] [PubMed] [Google Scholar]
- 51.Yamazaki H., Chan J., Ikura M., Michikawa T., Mikoshiba K. Tyr-167/Trp-168 in type 1/3 inositol 1,4,5-trisphosphate receptor mediates functional coupling between ligand binding and channel opening. J. Biol. Chem. 2010;285:36081–36091. doi: 10.1074/jbc.M110.140129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gangopadhyay J. P., Ikemoto N. Role of the Met3534–Ala4271 region of the ryanodine receptor in the regulation of Ca2+ release induced by calmodulin binding domain peptide. Biophys. J. 2006;90:2015–2026. doi: 10.1529/biophysj.105.074328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adkins C. E., Taylor C. W. Lateral inhibition of inositol 1,4,5-trisphosphate receptors by cytosolic Ca2+ Curr. Biol. 1999;9:1115–1118. doi: 10.1016/s0960-9822(99)80481-3. [DOI] [PubMed] [Google Scholar]
- 54.Veresov V. G., Konev S. V. Bridging the gaps in 3D structure of the inositol 1,4,5-trisphosphate-binding core. Biochem. Biophys. Res. Commun. 2006;341:1277–1285. doi: 10.1016/j.bbrc.2006.01.088. [DOI] [PubMed] [Google Scholar]
- 55.Bosanac I., Alattia J.-R., Mal T. K., Chan J., Talarico S., Tong F. K., Tong K. I., Yoshikawa F., Furuichi T., Iwai M., et al. Structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with its ligand. Nature. 2002;420:696–700. doi: 10.1038/nature01268. [DOI] [PubMed] [Google Scholar]
- 56.Lin C. C., Baek K., Lu Z. Apo and InsP3-bound crystal structures of the ligand-binding domain of an InsP3 receptor. Nat. Struct. Mol. Biol. 2011;18:1172–1174. doi: 10.1038/nsmb.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.