Abstract
Furfural is an inhibitory side product formed during the depolymerization of hemicellulose with mineral acids. In Escherichia coli, furfural tolerance can be increased by expressing the native fucO gene (encoding lactaldehyde oxidoreductase). This enzyme also catalyzes the NADH-dependent reduction of furfural to the less toxic alcohol. Saturation mutagenesis was combined with growth-based selection to isolate a mutated form of fucO that confers increased furfural tolerance. The mutation responsible, L7F, is located within the interfacial region of FucO homodimers, replacing the most abundant codon for leucine with the most abundant codon for phenylalanine. Plasmid expression of the mutant gene increased FucO activity by more than 10-fold compared to the wild-type fucO gene and doubled the rate of furfural metabolism during fermentation. No inclusion bodies were evident with either the native or the mutated gene. mRNA abundance for the wild-type and mutant fucO genes differed by less than 2-fold. The Km (furfural) for the mutant enzyme was 3-fold lower than that for the native enzyme, increasing efficiency at low substrate concentrations. The L7F mutation is located near the FucO N terminus, within the ribosomal binding region associated with translational initiation. Free-energy calculations for mRNA folding in this region (nucleotides −7 to +37) were weak for the native gene (−4.1 kcal mol−1) but weaker still for the fucO mutant (−1.0 to −0.1 kcal mol−1). The beneficial L7F mutation in FucO is proposed to increase furfural tolerance by improving gene expression and increasing enzyme effectiveness at low substrate levels.
INTRODUCTION
The enzyme l-1,2-propanediol oxidoreductase (encoded by fucO) is an NADH-linked, iron-dependent group III dehydrogenase. Typically, this enzyme functions only during the catabolism of deoxy sugars, where it catalyzes the reduction of lactaldehyde (1–3). FucO is a homodimer in which each subunit contains an active site (4). This enzyme has a broad substrate range (5) that includes furfural (6), a toxic side product in dilute acid hydrolysates of hemicellulose and an important inhibitor of microbial fermentations (7–9). Expression of fucO from plasmids has been used to improve furfural tolerance in Escherichia coli-based fermentations for ethanol and lactic acid (6). The reduction of furfural to the less toxic alcohol seems essential for growth and fermentation of dilute acid hydrolysates of hemicellulose (10–13).
Deoxy sugars such as fucose seldom dominate in the natural environment of E. coli (14). The fuc genes encoding deoxy sugar metabolism remain silent unless induced by fucose or other related sugars in the absence of competing substrates (1, 2). Although a natural product, furfural is unlikely to be an important natural substrate for this enzyme.
In this study, we have used site-specific mutagenesis and growth-based selection to identify a fucO mutation that confers a further increase in furfural tolerance.
MATERIALS AND METHODS
Strains, media, and genetic manipulations.
The strains, plasmids, and primers used in this study are listed in Table 1 (6, 15, 16). LB medium containing xylose was used for the construction of ethanol strains. AM1 minimal salts medium with xylose (17) was used for the maintenance and growth of ethanologenic strains. Solid medium contained 20 g liter−1 xylose. Broth cultures contained 50 g liter−1 xylose. Batch fermentations contained 100 g liter−1 xylose. Cultures were incubated at 37°C unless stated otherwise. Plates streaked with ethanologenic strains were incubated under argon.
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristic(s) or sequencea,b,c | Reference or source |
|---|---|---|
| Strains | ||
| TOP10F′ | F′ [lacIq Tn10(Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR nupG recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 λ− | Life Technologies |
| BL21λ(DE3) | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 Sam7 nin5] | Promega |
| XW92 | LY180 ΔyqhD | 16 |
| LY180a | ΔfrdBC::(frgZm celYEc) ΔldhA::(frgZm casABKo) adhE::(frgZm estZPp FRT) ΔackA::FRT rrlE::(pdc adhA adhB FRT) ΔmgsA::FRT | 15 |
| Plasmids | ||
| pCR2.1-TOPO | Plac, bla, kan | Life Technologies |
| pTrc99A | Ptrc, bla, lacIq | Pharmacia |
| pLOI4319 | fucO in pTrc99a | 6 |
| pET-15b | T7 expression vector | Novagen |
| pLOI5535 | fucO(L7FA47V) in pTrc99a | This study |
| pLOI5536 | fucO(L7F) in pTrc99a | This study |
| pLOI5537 | fucO(A47V) in pTrc99a | This study |
| pLOI5538 | fucO with His tag in pET15b | This study |
| pLOI5539 | fucO(L7F) with His tag in pET15b | This study |
| Primers | ||
| Thr143-lib1-For | CAATCCCCACCACAGCAGGCNNKGCGGCAGAAGTGACCATTAAC | This study |
| Thr143-lib1-Rev | CACGTAGTTAATGGTCACTTCTGCCGCMNNGCCTGCTGTGGTG | This study |
| Asn150-lib2-For | CTGCGGCAGAAGTGACCATTNNKTACGTGATCACTGACGAAGA | This study |
| Asn150-lib2-Rev | CTTCGTCAGTGATCACGTAMNNAATGGTCACTTCTGCCGCAGTG | This study |
| Val152-lib3-For | CAGAAGTGACCATTAACTACNNKATCACTGACGAAGAAAAAC | This study |
| Val152-lib3-Rev | GCGCCGTTTTTCTTCGTCAGTGATMNNGTAGTTAATGGTCAC | This study |
| Lys161-lib4-For | GACGAAGAAAAACGGCGCNNKTTTGTTTGCGTTGATCCGC | This study |
| Lys161-lib4-Rev | GATATCATGCGGATCAACGCAAACAAAMNNGCGCCGTTTTTC | This study |
| Val163-lib5-For | CGGCGCAAGTTTNNKTGCGTTGATCCGCATGATATCCCGCAGG | This study |
| Val163-lib5-Rev | CCTGCGGGATATCATGCGGATCAACGCAMNNAAACTTGCGCCG | This study |
| Phe253-lib6-For | GCAGTATGTTGCGGGTATGGGCNNKTCGAATGTTGGGTTAGG | This study |
| Phe253-lib6-Rev | CACCAACCCTAACCCAACATTCGAMNNGCCCATACCCGCAAC | This study |
| Val165-lib7-For | CGGCGCAAGTTTGTTTGCNNKGATCCGCATGATATCCCG | This study |
| Val165-lib7-Rev | CGGGATATCATGCGGATCMNNGCAAACAAACTTGCGCCG | This study |
| Thr206-lib8-For | GCTATTGAGGGGTATATTNNKCGTGGCGCGTGGGCGCTA | This study |
| Thr206-lib8-Rev | TAGCGCCCACGCGCCACGMNNAATATACCCCTCAATAGC | This study |
| Gly257-lib9-For | ATGGGCTTCTCGAATGTTNNKTTAGGGTTGGTGCATGGT | This study |
| Gly257-lib9-Rev | ACCATGCACCAACCCTAAMNNAACATTCGAGAAGCCCAT | This study |
| Cys361-lib10-For | GCGGCACTGGATGATGTTNNKACCGGTGGCAACCCGCGT | This study |
| Cys361-lib10-Rev | ACGCGGGTTGCCACCGGTMNNAACATCATCCAGTGCCGC | This study |
| Gly363-lib11-For | CTGGATGATGTTTGTACCNNKGGCAACCCGCGTGAAGCA | This study |
| Gly363-lib11-Rev | TGCTTCACGCGGGTTGCCMNNGGTACAAACATCATCCAG | This study |
| Gly364-lib12-For | GATGATGTTTGTACCGGTNNKAACCCGCGTGAAGCAACG | This study |
| Gly364-lib12-Rev | CGTTGCTTCACGCGGGTTMNNACCGGTACAAACATCATC | This study |
| Ile6-lib13-For | ATGATGGCTAACAGAATGNNKCTGAACGAAACGGCATGGTTTGG | This study |
| Ile6-lib13-Rev | CCAAACCATGCCGTTTCGTTCAGMNNCATTCTGTTAGCCATCAT | This study |
| Leu7-lib14-For | ATGATGGCTAACAGAATGATTNNKAACGAAACGGCATGGTTTGG | This study |
| Leu7-lib14-Rev | CCAAACCATGCCGTTTCGTTMNNAATCATTCTGTTAGCCATCAT | This study |
| F-L7F-fucO | GCTAACAGAATGATTTTTAACGAAACGGCATGGTTTGGT | This study |
| R-L7F-fucO | ACCAAACCATGCCGTTTCGTTAAAAATCATTCTGTTAGC | This study |
| F-A47V-fucO | CTGGTGCAATGCGGCGTGGTGGTGAAAGTGACCGATAAGATGG | This study |
| R-A47V-fucO | CCATCTTATCGGTCACTTTCACCACCACGCCGCATTGCACCAG | This study |
| fucO-F-EcoRI | CCATGGAATTCGATTGCCGTAGTGCTG | This study |
| fucO-R-BamHI | GCGGATCCTGCGGTTGGTACGGTAACGGCGA | This study |
| fucO-F-6His-NcoI | CATGCCATGGCGATGGCTAACAGAATG | This study |
| fucO-R-6His-BamHI | GCGGGATCCTTAATGATGATGATGATGATGCCAGGCGGTATGGTAAAGCTCTACAATAT | This study |
| bla-RT-F | GCTATGTGGCGCGGTATTAT | This study |
| bla-RT-R | AAGTAAGTTGGCCGCAGTGT | This study |
| fucO-RT-F | ACGCCGTGGTTATCAGAAGG | This study |
| fucO-RT-R | CAGGTAATCCGCGCCGCTAT | This study |
LY180 is an ethanologenic derivative of E. coli W containing integrated genes from other organisms and various chromosomal mutations. Note that the celY gene (endoglucanase) from Erwinia carotovora is expressed from a Zymomonas mobilis surrogate promoter and integrated into the frd region. The Klebsiella oxytoca casAB operon is expressed from a Z. mobilis surrogate promoter and integrated into the ldhA region. The Z. mobilis pdc adhA adhB artificial operon for ethanol production is integrated and expressed from the native rrlE promoter. The Pseudomonas putida estZ (acetyl esterase) is integrated into adhE and expressed from a surrogate promoter from Z. mobilis.
Restriction enzyme sites used in construction are underlined.
NNK and MNN were used as codon degeneracy in forward and reverse primers, respectively.
Standard genetic methods were used for the isolation of DNA and plasmids, digestion with restriction enzymes, PCR amplification of DNA, and plasmid constructions (18). Enzymes were purchased from New England BioLabs (Ipswich, MA) and used as directed by the vendor. Plasmid constructions were confirmed by Sanger sequencing.
Design and construction of fucO libraries.
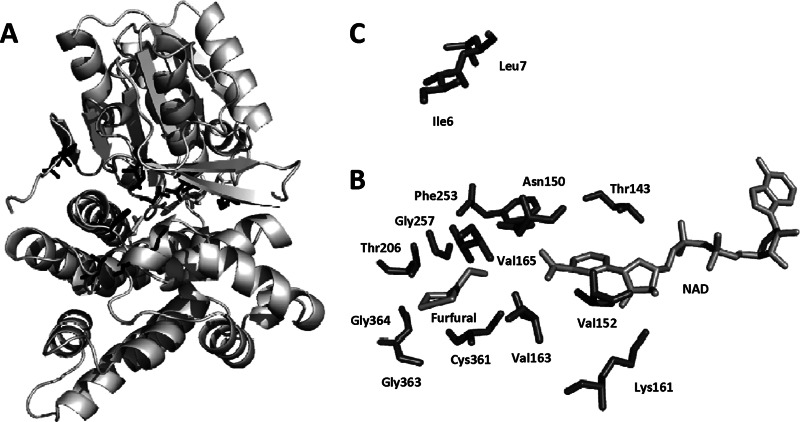
AutoDock (19) was used to position furfural into the active site of FucO (4). This docked enzyme model served as a guide for library constructions (Fig. 1A). The first group of libraries was designed for the 5-Å region near furfural (designated Lib1 T143NNK, Lib2 N150NNK, Lib3 V152NNK, Lib4 K161NNK, Lib5 V163NNK, Lib6 F253NNK, Lib7 V165NNK, Lib8 T206NNK, Lib9 G257NNK, Lib10 C361NNK, Lib11 G363NNK, and Lib12 G364NNK). A second group of libraries was designed for the interface region of the homodimers (designated Lib13 I6NNK and Lib14 L7NNK). All 14 single-residue libraries were constructed by using QuikChange EZ (Agilent Technologies, Santa Clara, CA) with plasmid pLOI4319 as the template. pLOI4319 (pTrc99A derivative) contained a 1,302-bp fragment downstream from the trc promoter consisting of the 3′ end of fucA, the intergenic sequence, the complete fucO ribosomal binding and coding regions, and an additional 71 bp downstream of the native transcriptional terminator. Following PCR, template DNA was digested with DpnI, leaving only the PCR product. Resulting plasmids were transformed into E. coli TOP10F′ (Life Technologies, Grand Island, NY) by electroporation. Plasmid libraries of more than 1,000 colonies were prepared for each mutated amino acid.
Fig 1.
Crystal structure of FucO. (A) The complete FucO structure. Twelve amino acid residues residing around the active site (B) and two residues at the homodimer interface (C) were targeted for mutagenesis. Libraries: Lib1 Thr143, Lib2 Asn150, Lib3 Val152, Lib4 Lys161, Lib5 Val163, Lib6 Phe253, Lib7 Val165, Lib8 Thr206, Lib9 Gly 257, Lib10 Cys361, Lib11 Gly363, Lib12 Gly364, Lib13 Ile6, and Lib14 Leu7.
Growth-based screening procedures.
Libraries were transformed into strain XW92 and spread onto AM1 plates containing xylose and ampicillin (100 μg ml−1). A total of 100 colonies from each library were screened individually. Colonies were inoculated into 100-well plates for seed culture growth (AM1 plus 5% xylose, 50 μg ml−1 ampicillin, and 0.025 mM isopropyl-β-d-thiogalactopyranoside [IPTG]) with the Bioscreen C growth curve analyzer (Growth Curves USA, Piscataway, NJ). Stationary-phase seed cultures (15 μl) were diluted into wells containing 0.39 ml screening medium (AM1 plus 10% xylose, 12.5 μg ml−1 ampicillin, 0.025 mM IPTG, and 12.5 mM furfural). Bioscreen C plates were incubated at 37°C (with shaking for 10 s at 30-min intervals). Cell density was recorded automatically. Beneficial mutations were identified by improved growth. These were confirmed by back transformation, subcloning, retesting, and sequencing.
Tube assays for furan tolerance (MIC) and batch fermentations.
Furfural toxicity was examined at 37°C by using culture tubes (13 by 100 mm) containing 4 ml of AM1 minimal salts medium (50 g liter−1 xylose, 12.5 μg liter−1 ampicillin, 0.025 mM IPTG, and furfural). Tube cultures were inoculated to an initial density of 43 mg (dry cell weight) liter−1. Conditions for MIC determinations and batch fermentation procedures were as previously described (15). Furfural was measured with a Beckman Coulter DU 800 spectrophotometer (20).
Assay of FucO activity and SDS-PAGE analysis.
Cell cultivation was as described for tube assays of furfural tolerance. Cells were harvested after 24 h. FucO activity was determined as previously described (6). One unit of activity is defined as the amount of enzyme that converts 1 μmol of NADH to NAD+ per min (6).
The protein concentrations in cell extracts and purified enzyme preparations were determined with the Pierce BCA protein reagent (Thermo, Waltham, MA) with bovine serum albumin as the standard. Whole-cell proteins and soluble proteins were examined by 12% SDS-PAGE (Bio-Rad, Hercules, CA).
Construction and purification of His-tagged FucO.
Wild-type FucO and the L7F mutant were purified by affinity chromatography. A tag of six histidine residues was added at the 3′ end of each gene. PCR products with this tag were ligated into pET15b and transformed into BL21λ(DE3). The resulting clones were designated pLOI5538 (wild type) and pLOI5539 (L7F). Expression and purification of recombinant FucO were done as previously described (6), except that the induction temperature was reduced to 30°C and 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) was included as a reducing reagent during purification. An Amicon ultra-15 centrifugal filter was used to remove imidazole and concentrate enzymes prior to kinetic studies. Since FucO is oxygen sensitive (21), cytoplasmic activities were notably more stable in the presence of TCEP.
Transcriptional analyses of mutant fucO(L7F).
The transcript abundance of native fucO was compared with that of the mutant fucO(L7F) gene by using cultures grown as described for tube assays of tolerance. Cells were harvested at 24 h by centrifugation at 4°C and washed once with cold distilled water. RNA was extracted and purified using RNeasy Mini columns (Qiagen) and digestion with DNase I. cDNA was prepared from 50 ng total RNA using SuperScript II. Samples were analyzed by using a Bio-Rad iCycler with SYBR green reverse transcription-PCR. Transcript abundance was estimated by using the bla gene (vector) as an internal standard (22, 23).
mRNA folding and energy calculations.
The minimum free energy of mRNA secondary structures was calculated for partial sequences of wild-type fucO and mutant fucO(L7F) by using the UNAFOLD web server (http://mfold.rna.albany.edu; 28) with the default parameters. The sequence window was set from nucleotide (nt) −7 through nt +37 with the A in ATG (start codon) being designated nt 0, as proposed by G. Kudla et al. (24). Energies were calculated for a series of seven segments in which the intergenic region was increased by increments of 10 bp.
RESULTS
FucO mutation L7F increases furfural tolerance.
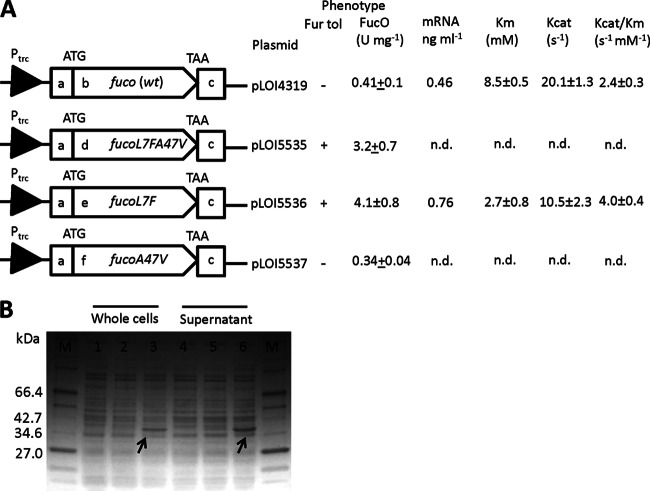
Two groups of libraries were designed on the basis of the X-ray structures of FucO and furfural (Fig. 1A) (4, 25). The first group included 12 libraries targeting amino acids associated with furfural docking (Fig. 1B). The second group targeted amino acids located at the interface between FucO homodimers (Fig. 1C). After all 14 libraries were tested for improvements in growth with 12.5 mM furfural, a single clone was recovered and designated pLOI5535. Sequencing revealed that the fucO gene in this plasmid contained two mutations, L7F from library 14 (homodimer interface) and an undirected PCR error, A47V. Site-specific mutagenesis was used to create corresponding single mutations in FucO, designated pLOI5536 and pLOI5537, respectively (Fig. 2A). Only plasmids that contained the L7F mutation (pLOI5535 and pLOI5536) improved growth in medium containing 12.5 mM furfural (Bioscreen C; see Fig. S1 in the supplemental material). All enhancement of furfural tolerance resided with the single L7F mutation, a conservative replacement of leucine with another hydrophobic amino acid (phenylalanine). The 10 amino acid residues at the N terminus are required for dimerization and essential for activity (4). Dimerization is dominated by hydrophobic interactions which should be retained by the replacement of leucine with phenylalanine.
Fig 2.
(A) Schematic representation plasmid constructs. Plasmid pLOI4319 contains the wild-type fucO gene. Plasmid pLOI5535 contains the original double mutant gene [fucO(L7FA47V)]. Plasmid pLOI5536 contains the fucO(L7F) gene. Plasmid pLOI5537 contains the fucO(A47V) gene. Assumed native ribosome binding site regions before ATG and the native terminator sequence after TAA were included while amplifying the fucO gene from E. coli W. Ptrc, the promoter of vector pTrc99a. Segments: a, the 82 bp of chromosome sequence before starting codon ATG of the fucO gene; b, the wild-type fucO gene; c, the 71 bp of chromosome sequence after stop codon TAA of the fucO gene; d, the double mutant fucO(L7FA47V) gene; e, the mutant fucO(L7F) gene; f, the mutant fucO(A47V) gene. All cultures were grown under MIC conditions as described in Materials and Methods. Cells were harvested after 24 h. Samples used for transcriptional analysis and FucO assay were prepared as described in Materials and Methods. “Fur tol” means the furfural tolerance of the recombinant strain with the corresponding plasmid in the presence of 12.5 mM furfural. (B) SDS-PAGE of crude extract from strains XW92 and pTrc99a (lanes 1 and 4) and pTrc99a derivatives containing fucO genes. Plasmid pLOI4319 containing the wild-type fucO gene (lanes 2 and 5). Plasmid pLOI5536 containing the mutated fucO(L7F) gene (lanes 3 and 6). Lanes 1, 2, and 3 contained the indicated whole-cell samples at 24 h. Lanes 4, 5, and 6 contained the indicated supernatant samples at 24 h. The arrows indicate the putative FucO protein from plasmid expression. M, molecular mass marker lanes. n.d., not determined.
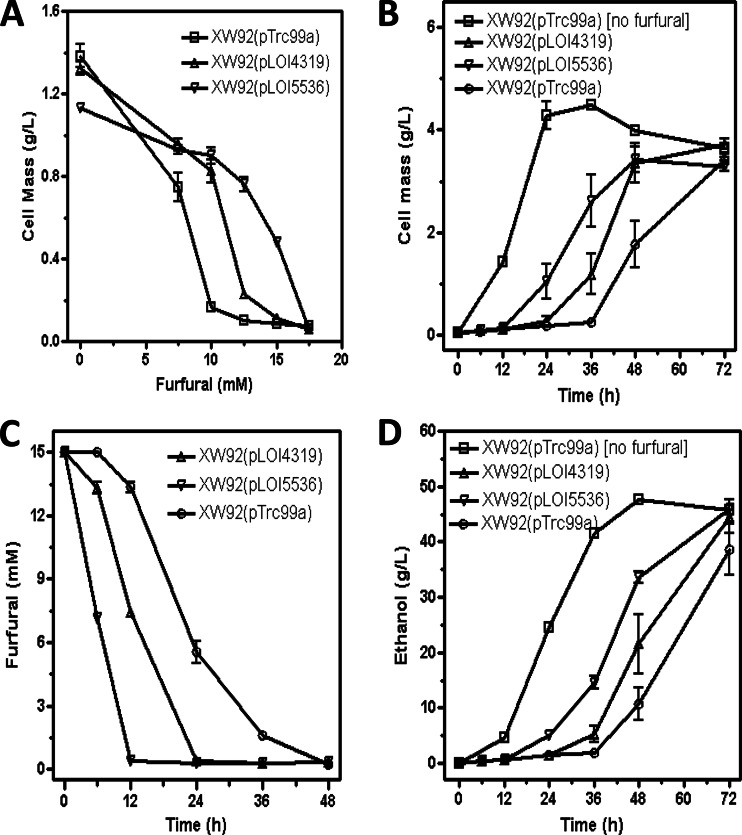
The L7F FucO mutation in strain XW92(pLOI5536) increased the MIC of furfural, in comparison to the wild-type gene (pLOI4319) or the empty vector (Fig. 3A). The L7F FucO mutation also improved the fermentation performance (strain XW92) in AM1 medium with 100 g liter−1 xylose and 15 mM furfural (Fig. 3B to D). With the mutant gene (pLOI5536), 15 mM furfural was completely metabolized in 12 h, compared to 24 h with the native fucO gene (pLOI4319) and 48 h with the empty vector. With all three strains, growth and ethanol production were delayed until furfural had been substantially metabolized to the corresponding alcohol.
Fig 3.
Increased expression of fucO increased the MIC of furfural and the furfural degradation rate in fermentation. (A) MIC of furfural. (B) Cell mass of fermentation on 15 mM furfural. (C) Furfural degradation of strains during fermentation with 15 mM furfural. (D) Ethanol production of strains during fermentation with 15 mM furfural.
Increased furfural reductase activity is responsible for improvement of furfural tolerance.
The furfural reductase activity (Fig. 2A) of strain XW92 harboring the L7F mutant fucO gene (pLOI5535 and pLOI5536) was 10-fold higher than that of XW92 harboring the wild-type or fucO(A47V) mutant gene (pLOI4319 or pLOI5537). SDS-PAGE (12.5% acrylamide) was used to compare the protein profiles (soluble and whole cell) of the fucO mutant strains (Fig. 2B). A new dense band was observed in the FucO region (38.2 kDa) of the L7F mutant (pLOI5536). This band was estimated to have >10 times the intensity of a corresponding band in XW92(pLOI4319) containing the wild-type gene and was absent from strains harboring the empty vector. His-tagged plasmid constructs with wild-type FucO and L7F mutant FucO were purified and characterized. The Km of L7F was 2.7 mM, 1/3 that of wild-type FucO (Fig. 2A). Thus, the increase in furfural tolerance associated with the L7F mutation in fucO appears to result from an increase in activity and an increase in enzyme effectiveness at low substrate concentrations.
The L7F mutation in fucO increases cytoplasmic FucO activity.
Changes in transcription, translation, and enzyme stability were investigated as potential causes of the large increase in cytoplasmic FucO in the L7F mutant. FucO mRNA levels were less than 2-fold higher in XW92(pLOI5536) containing the mutant gene than in XW92(pLOI4319) containing the wild-type gene (Fig. 2A). This may contribute to increased expression but is insufficient to explain the 10-fold increase in FucO activity (Fig. 2A). Enzyme stability was also investigated by comparing activity in stationary-phase cells incubated at 37°C. After 96 h, wild-type furfural reductase activity in whole cells of XW92(pLOI4319) declined by 10% ± 9%, while that of the L7F mutant (pLOI5536) had declined by 20% ± 2%, eliminating differential protein stability as a basis for the increase in cytoplasmic FucO.
Folded mRNA structures have been shown to reduce protein synthesis by slowing translation or translational initiation (26, 27). The L7F mutation is near the N terminus of FucO and within the ribosomal binding region. The genetic region corresponding to this N terminus encodes a segment of protein essential for both quaternary structure and activity (4). Although the L7F mutation represents a conservative amino acid replacement and did not alter specific activity, the nucleotide changes could have a more dramatic effect on mRNA folding and translation.
In E. coli, the fucO gene is downstream from fucA and is part of the fucA-fucO operon. Using the UNAFOLD Web server (http://mfold.rna.albany.edu/; 28), minimal energy structures were predicted for a series of seven sequence segments that span the region from 38 bases of the fucO 5′ end through the 27-base intergenic region (segments 1 to 3) and into the fucA 3′ end (segments 4 to 7), increasing in length by 10-base increments. Calculated ΔG energies for all segments tested and all predicted structures for the wild-type gene were more stable (lower ΔG) than corresponding regions of the fucO(L7F) mutant (Table 2). The largest difference was observed for the shortest segment examined, the 5′ end of fucO plus 7 upstream bases. A single stem was predicted for this segment (4.3 kcal mol−1), and this stem includes the codon (CUG) at amino acid residue 7. The mutation at position 7 to UUU eliminated this structure and was predicted to form a weaker structure. Weak mRNA structures in this region (−7 to +27 region) have been shown to correlate with increased translational initiation and improved expression (26, 29). Although predicted structures for both the native and mutant fucO segments were not strong, the mutant structures were consistently weaker.
Table 2.
Predicted mRNA folding energies
| mRNAa segment, region | Folding energy (kcal mol−1)b |
Difference in energy (kcal mol−1)c | |
|---|---|---|---|
| Wild-type fucO gene | Mutant fucO(L7F) gene | ||
| 1, −7 to +37 | −4.3 (1) | −0.38 ± 0.41 (6) | +3.92 |
| 2, −17 to +37 | −4.2 ± 0.46 (3) | −2.85 ± 0.64 (2) | +1.35 |
| 3, −27 to +37 | −8.0 (1) | −4.43 ± 0.42 (4) | +3.67 |
| 4, −37 to +37 | −10.9 (1) | −7.05 ± 0.35 (2) | +3.85 |
| 5, −47 to +37 | −10.33 ± 0.51 (3) | −8.47 ± 0.77 (3) | +1.86 |
| 6, −57 to +37 | −13.48 ± 0.45 (5) | −12.0 ± 0.53 (3) | +1.48 |
| 7, −67 to +37 | −15.84 ± 0.38 (5) | −14.0 ± 0.53 (3) | +1.84 |
Segments were numbered by designating the adenosine nucleotide of the fucO start codon zero, increasing in the direction of translation. Negative numbers from −1 to −27 represent bases in the intergenic region between fucA and fucO (upstream) and proceeding further upstream into the 3′ end of fucA (−27 to −67).
Minimal folding energies and structures were predicted by the UNAFOLD web server (http://mfold.rna.albany.edu; 28). Average values are presented with standard deviations. The values in parentheses are the numbers of predicted structures.
Wild-type fucO gene minus mutant fucO(L7F) gene. Energies for all predicted structures were more positive (weaker) for the mutant fucO(L7F) than for the wild-type mRNA segments.
The 10-fold increase in cytoplasmic activity caused by the L7F mutation appears to result from improved translation rather than increased transcription or changes in mRNA or protein stability.
DISCUSSION
The crystal structure of FucO has been described previously and exhibits remarkable similarity to two other oxidoreductases in E. coli, YqhD (NADPH-linked furfural reductase) (30, 31) and AdhE (NADH-linked alcohol dehydrogenase) (32, 40). All three of these enzymes have broad substrate ranges. Although YqhD can effectively reduce furfural, the utility of this enzyme is severely limited by the low Km for NADPH that competes with essential biosynthetic reactions (15, 33). NADH is abundant during fermentative metabolism, and the small amounts needed for furfural reduction by fucO should not adversely impact growth or product yields.
Removal of furfural from lignocellulosic sugars is essential for effective fermentation (10, 39). The toxicity of hydrolysates is correlated with the concentration of furfural (9, 13). Cells typically initiate rapid growth only after furfural is fully metabolized (10). Plasmid-based expression of fucO improved furfural tolerance (6) and provided an opportunity for further improvement by site-specific mutagenesis. Growth-based selection for furfural tolerance provided a powerful screening method, recovering a single mutation (L7F) that increased cytoplasmic activity 10-fold and increased the affinity for furfural (3-fold lower Km).
Many methods are available to create sequence diversity libraries, including chemical mutagenesis (34), error-prone PCR, saturation mutagenesis, and DNA shuffling (29, 35, 36, 41). However, effective screening often remains as a bottleneck. Recently, a Q263R transaldolase mutant was obtained by directed evolution with a 5-fold increase in activity. Almost 60,000 colonies were subjected to high-throughput TAL activity screening with a sensitive fluorescence assay (37). A similar success (site-specific mutagenesis and growth-based screening) has been reported previously for the expression of Piromyces sp. xylose isomerase (xylA) in Saccharomyces cerevisiae (38). Cytoplasmic activities were increased by 77%, and ethanol production from xylose was dramatically improved (38). In contrast, 14 single residues were targeted in FucO for saturation mutagenesis libraries based on a visual inspection of furfural docked in the FucO structure (Fig. 1). The fucO(L7F) mutant exhibited a 10-fold increase in cytoplasmic activity and was isolated after the screening of only 1,400 colonies. The site of this mutation was unexpected, with the contact region of homodimers distant from the active site. With the mutant gene, furfural was metabolized in vivo at twice the rate of the native enzyme during fermentation.
Supplementary Material
ACKNOWLEDGMENTS
L. O. Ingram is a consultant for the Myriant Corporation and is a minor shareholder.
This research was supported by the Myriant Corporation, the Department of Agriculture (grant 2011-10006-30358), and the Department of Energy (grant DE-PI0000031).
Footnotes
Published ahead of print 8 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00149-13.
REFERENCES
- 1. Boronat A, Aguilar J. 1979. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 140:320–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen YM, Lin ECC. 1984. Dual control of a common l-1,2-propanediol oxidoreductase by l-fucose and l-rhamnose in Escherichia coli. J. Bacteriol. 157:828–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu Y, Lin ECC. 1989. l-1,2-Propanediol exits more rapidly than l-lactaldehyde from Escherichia coli. J. Bacteriol. 171:862–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Montella C, Bellsolell L, Perez-Luque R, Badia J, Baldoma L, Coll M, Aguilar J. 2005. Crystal structure of an iron-dependent group III dehydrogenase that interconverts l-lactaldehyde and l-1,2-propanediol in Escherichia coli. J. Bacteriol. 187:4957–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blikstad C, Widersten M. 2010. Functional characterization of a stereospecific diol dehydrogenase, FucO, from Escherichia coli: substrate specificity, pH dependence, kinetic isotope effects and influence of solvent viscosity. J. Mol. Catal. B Enzym. 66:148–155 [Google Scholar]
- 6. Wang X, Miller EN, Yomano LP, Zhang X, Shanmugam KT, Ingram LO. 2011. Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl. Environ. Microbiol. 77:5132–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mills TY, Sandoval NR, Gill RT. 2009. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol. Biofuels 2:11 doi:10.1186/1754-6834-2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parawira W, Tekere M. 2011. Biotechnological strategies to overcome inhibitors in lignocellulose hydrolysates for ethanol production: review. Crit. Rev. Biotechnol. 31:20–31 [DOI] [PubMed] [Google Scholar]
- 9. Zheng HB, Wang X, Yomano LP, Shanmugam KT, Ingram LO. 2012. Increase in furfural tolerance in ethanologenic Escherichia coli LY180 by plasmid-based expression of thyA. Appl. Environ. Microbiol. 78:4346–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Geddes CC, Nieves IU, Ingram LO. 2011. Advances in ethanol production. Curr. Opin. Biotechnol. 22:312–319 [DOI] [PubMed] [Google Scholar]
- 11. Martinez A, Rodriguez ME, Wells ML, York SW, Preston JF, Ingram LO. 2001. Detoxification of dilute acid hydrolysates of lignocellulose with lime. Biotechnol. Prog. 17:287–293 [DOI] [PubMed] [Google Scholar]
- 12. Martinez A, Rodriguez ME, York SW, Preston JF, Ingram LO. 2000. Effects of Ca(OH)2 treatments (“overliming”) on the composition and toxicity of bagasse hemicellulose hydrolysates. Biotechnol. Bioeng. 69:526–536 [DOI] [PubMed] [Google Scholar]
- 13. Zaldivar J, Martinez A, Ingram LO. 1999. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 65:24–33 [DOI] [PubMed] [Google Scholar]
- 14. Mayer C, Boos W. 29 March 2005, posting date Chapter 3.4.1, Hexose/pentose and hexitol/pentitol metabolism. In Böck A. (ed), EcoSal---Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: http://www.ecosal.org [Google Scholar]
- 15. Miller EN, Jarboe LR, Yomano LP, York SW, Shanmugam KT, Ingram LO. 2009. Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl. Environ. Microbiol. 75:4315–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Yomano LP, Lee JY, York SW, Zheng HB, Mullinnix MT, Shanmugam KT, Ingram LO. 2013. Engineering furfural tolerance in Escherichia coli improves the fermentation of lignocellulosic sugars into renewable chemicals. Proc. Natl. Acad. Sci. U. S. A. 110:4021–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martinez A, Grabar TB, Shanmugam KT, Yomano LP, York SW, Ingram LO. 2007. Low salt medium for lactate and ethanol production by recombinant Escherichia coli B. Biotechnol. Lett. 29:397–404 [DOI] [PubMed] [Google Scholar]
- 18. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30:2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martinez A, Rodriguez ME, York SW, Preston JF, Ingram LO. 2000. Use of UV absorbance to monitor furans in dilute acid hydrolysates of biomass. Biotechnol. Prog. 16:637–641 [DOI] [PubMed] [Google Scholar]
- 21. Cabiscol E, Hidalgo E, Badia J, Baldoma L, Ros JQ, Aguilar J. 1990. Oxygen regulation of l-1,2-propanediol oxidoreductase activity in Escherichia coli. J. Bacteriol. 172:5514–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Su Y, Rhee MS, Ingram LO, Shanmugam KT. 2011. Physiological and fermentation properties of Bacillus coagulans and a mutant lacking fermentative lactate dehydrogenase activity. J. Ind. Microbiol. Biotechnol. 38:441–450 [DOI] [PubMed] [Google Scholar]
- 23. Wang QZ, Ingram LO, Shanmugam KT. 2011. Evolution of d-lactate dehydrogenase activity from glycerol dehydrogenase and its utility for d-lactate production from lignocellulose. Proc. Natl. Acad. Sci. U. S. A. 108:18920–18925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kudla G, Murray AW, Tollervey D, Plotkin JB. 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller EN, Jarboe LR, Turner PC, Pharkya P, Yomano LP, York SW, Nunn D, Shanmugam KT, Ingram LO. 2009. Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl. Environ. Microbiol. 75:6132–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jia M, Li Y. 2005. The Relationship among gene expression, folding free energy and codon usage bias in Escherichia coli. FEBS Lett. 579:5333–5337 [DOI] [PubMed] [Google Scholar]
- 27. Tsao D, Shabalina SA, Gauthier J, Dokholyan NV, Diatchenko L. 2011. Disruptive mRNA folding increases translational efficiency of catechol-O-methyltransferase variant. Nucleic Acids Res. 39:6201–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Labrou NE. 2010. Random mutagenesis methods for in vitro directed enzyme evolution. Curr. Protein Pept. Sci. 11:91–100 [DOI] [PubMed] [Google Scholar]
- 30. Jarboe LR. 2011. YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl. Microbiol. Biotechnol. 89:249–257 [DOI] [PubMed] [Google Scholar]
- 31. Sulzenbacher G, Alvarez K, van den Heuvel RHH, Versluis C, Spinelli M, Campanacci V, Valencia C, Cambillau C, Eklund H, Tegoni M. 2004. Crystal structure of E. coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J. Mol. Biol. 342:489–502 [DOI] [PubMed] [Google Scholar]
- 32. Conway T, Ingram LO. 1989. Similarity of Escherichia coli propanediol oxidoreductase (fucO product) and an unusual alcohol dehydrogenase from Zymomonas mobilis and Saccharomyces cerevisiae. J. Bacteriol. 171:3754–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Turner PC, Miller EN, Jarboe LR, Baggett CL, Shanmugam KT, Ingram LO. 2011. YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance. J. Ind. Microbiol. Biotechnol. 38:431–439 [DOI] [PubMed] [Google Scholar]
- 34. Kim Y, Ingram LO, Shanmugam KT. 2008. Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. J. Bacteriol. 190:3851–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reetz MT. 2011. Laboratory evolution of stereoselective enzymes: a prolific source of catalysts for asymmetric reactions. Angew. Chem. Int. Ed. Engl. 50:138–174 [DOI] [PubMed] [Google Scholar]
- 36. Wang M, Si T, Zhao HM. 2012. Biocatalyst development by directed evolution. Bioresour. Technol. 115:117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen SH, Hwang DR, Chen GH, Hsu NS, Wu YT, Li TL, Wong CH. 2012. Engineering transaldolase in Pichia stipitis to improve bioethanol production. ACS Chem. Biol. 7:481–486 [DOI] [PubMed] [Google Scholar]
- 38. Lee S-M, Jellison T, Alper HS. 2012. Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 78:5708–5716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chundawat SPS, Vismeh R, Sharma LN, Humpula JF, Sousa LD, Chambliss CK, Jones AD, Balan V, Dale BE. 2010. Multifaceted characterization of cell wall decomposition products formed during ammonia fiber expansion (AFEX) and dilute acid based pretreatments. Bioresour. Technol. 101:8429–8438 [DOI] [PubMed] [Google Scholar]
- 40. Reid MF, Fewson CA. 1994. Molecular characterization of microbial alcohol dehydrogenases. Crit. Rev. Microbiol. 20:13–56 [DOI] [PubMed] [Google Scholar]
- 41. Roodveldt C, Aharoni A, Tawfik DS. 2005. Directed evolution of proteins for heterologous expression and stability. Curr. Opin. Struct. Biol. 15:50–56 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.