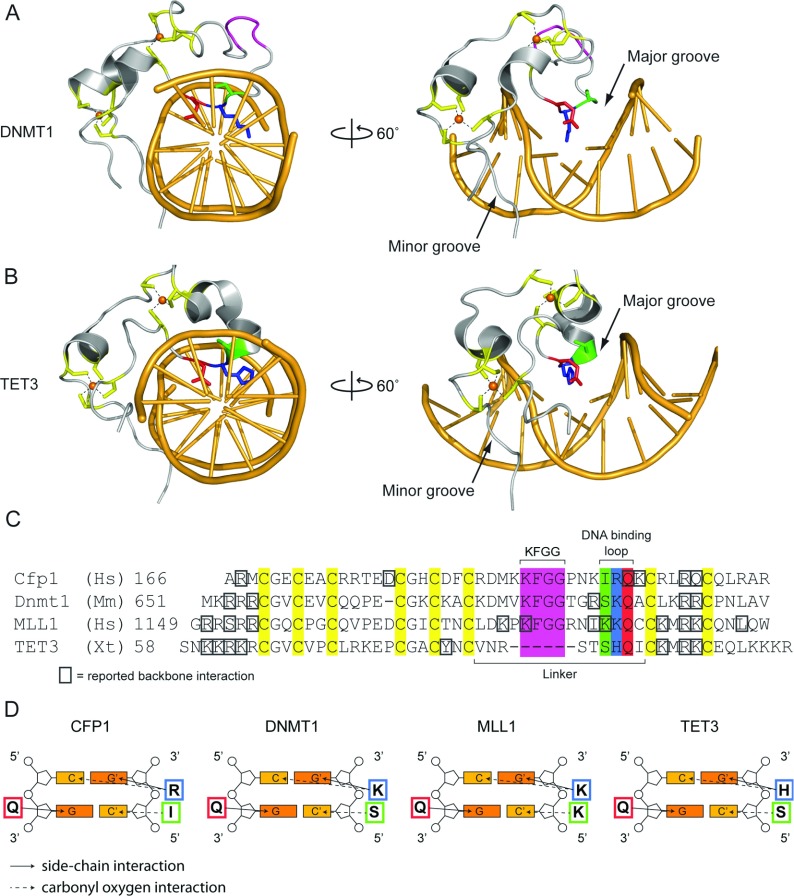
Figure 3. Structural insight into the DNA-binding properties of ZF-CxxC proteins.
(A and B) Crystal structures of the (A) Homo sapiens DNMT1 (PDB code 3PTA) and (B) Xenopus tropicalis TET3 (PDB code 4HP3) ZF-CxxC domains in complex with DNA, viewed down the double-helix axis (left) and rotated 60° to the right (right). For both structures, the eight cysteine residues are highlighted in yellow and their interaction with the Zn2+ ions (represented as orange spheres) are shown by dashes. The KFGG motif is highlighted in pink and the DNA-binding tripeptide is highlighted in green, blue and red for serine, lysine/histidine and glutamine respectively. The right-hand panels highlight that the DNA-binding tripeptide loop interrogates the CpG dinucleotide via the major groove of DNA, and that the N- and C-terminal parts of the ZF-CxxC domain interact with the minor groove. Zn2+ ions are represented as spheres. (C) A manually curated multiple sequence alignment of the amino acid sequence of four ZF-CxxC structures (PDB codes: CFP1, 3QMG; DNMT1, 3PTA; MLL, 2KKF; TET3, 4HP3). Residues reported to interact with the DNA backbone are marked with a grey box. Other residues are highlighted as in (A) and (B). (D) Schematic representations of the base-specific hydrogen bond interactions between the DNA-binding tripeptide and a CpG dinucleotide. Side-chain interactions are shown by a continuous arrow and carbonyl oxygen interactions are shown by a broken arrow.