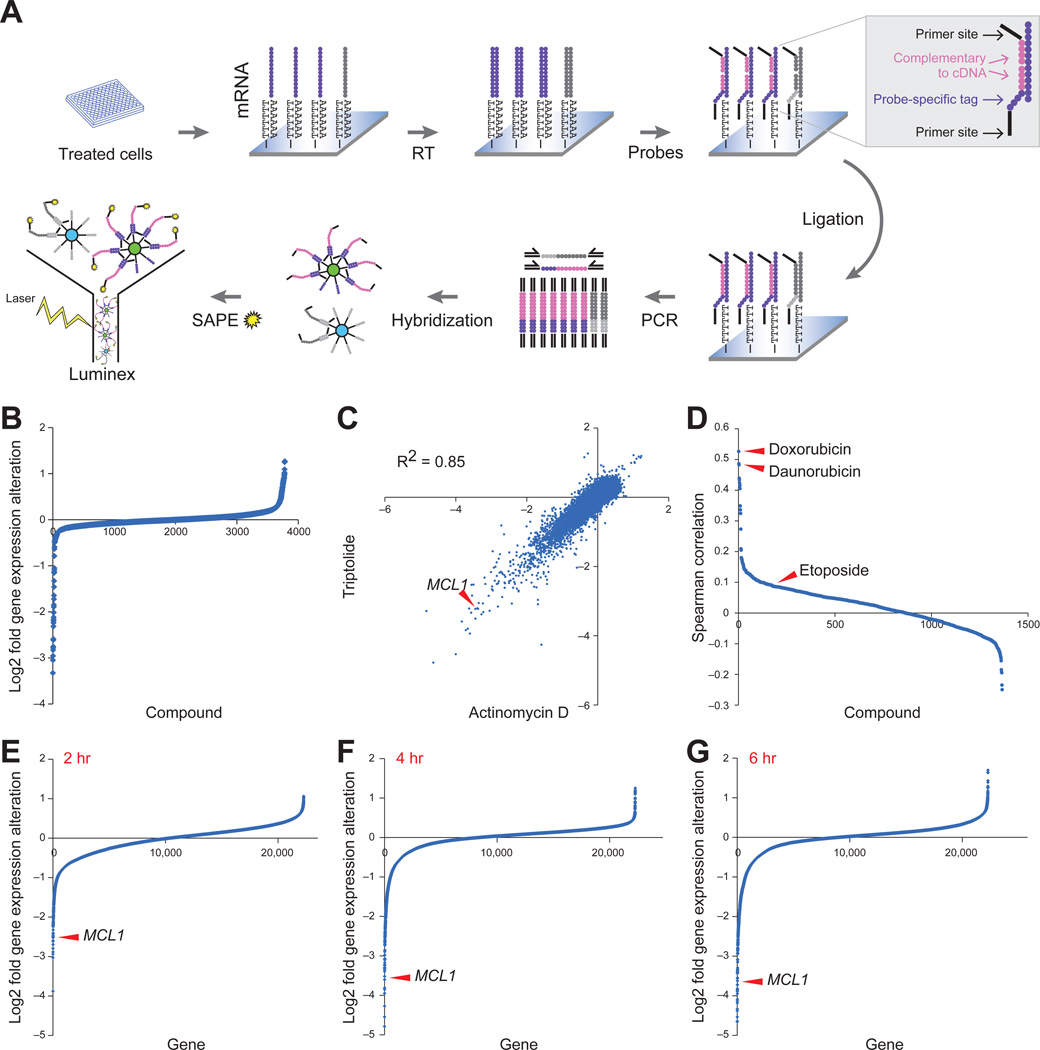
Figure 1. Bead-based high-throughput gene expression screening identified MCL1 repression by transcriptional inhibitor compounds.
A. Illustration of screening procedure. mRNA levels of MCL1 and 48 other apoptotic genes were measured in MCF7 cells 8 hours after treatment with 2,922 small molecules.
B. MCL1 expression modulation by 2,922 compounds. Compounds and DMSO controls were sorted by MCL1 expression repression. The y-axis displays log2 gene expression fold change.
C. Gene expression profiling by Affymetrix microarrays of MCF7 cells with triptolide (500 nM) and actinomycin D (2.5 µM) for 4 hours. Both the x- and y-axis displays log2 gene expression fold alteration.
D. The 1,317 compounds used to treat MCF7 cells in the Connectivity Map were displayed in descending order of their correlation with triptolide (calculated as the Spearman rank correlation of the differential gene expression across the Affymetrix U133A chip).
E-G. Gene expression repression by triptolide at 2 hours (E), 4 hours (F) and 6 hours (G). The genes were ranked by the extent of repression. The y-axis displays log2 gene expression fold alteration. Arrowheads indicate MCL1.
See also Figure S1 and Tables S1 and S2.