Abstract
According to multi-level theory, evolutionary transitions require mediating conflicts between lower-level units in favour of the higher-level unit. By this view, the origin of eukaryotes and the origin of multicellularity would seem largely equivalent. Yet, eukaryotes evolved only once in the history of life, whereas multicellular eukaryotes have evolved many times. Examining conflicts between evolutionary units and mechanisms that mediate these conflicts can illuminate these differences. Energy-converting endosymbionts that allow eukaryotes to transcend surface-to-volume constraints also can allocate energy into their own selfish replication. This principal conflict in the origin of eukaryotes can be mediated by genetic or energetic mechanisms. Genome transfer diminishes the heritable variation of the symbiont, but requires the de novo evolution of the protein-import apparatus and was opposed by selection for selfish symbionts. By contrast, metabolic signalling is a shared primitive feature of all cells. Redox state of the cytosol is an emergent feature that cannot be subverted by an individual symbiont. Hypothetical scenarios illustrate how metabolic regulation may have mediated the conflicts inherent at different stages in the origin of eukaryotes. Aspects of metabolic regulation may have subsequently been coopted from within-cell to between-cell pathways, allowing multicellularity to emerge repeatedly.
Keywords: endosymbiosis, genome transfer, levels of selection, major transitions, mitochondria, redox signalling
Regardless of how early eukaryotes escaped from their predicament, it is plain that the problems faced by a prokaryotic host cell with bacterial endosymbionts are serious, if not irreconcilable, and go a long way towards explaining why there are no surviving evolutionary intermediates between prokaryotes and eukaryotes [1, p. 13].
1. Introduction
In a groundbreaking synthesis of genomics and bioenergetics, Lane & Martin [2] examine the question: Why have not prokaryotes evolved complex multicellularity? Their analysis points out crucial differences between prokaryotes and eukaryotes in the availability of energy and the role of energetic differences in the capacity for gene expression. By their view, the mitochondrial endosymbiosis coupled with the accumulation of mitochondrial genes in the nuclear genome allowed tight regulation of respiration at relatively low cost in terms of genome size. This genetic and bioenergetic complexity allows eukaryotes to then easily evolve complex multicellularity. Left unaddressed by their analysis is an obvious follow-up question: if eukaryotes are so richly favoured, why did they only evolve once?
Metabolic complementation could have produced a eukaryotic cell in a variety of ways from a variety of partners [3], yet the data suggest that eukaryotes are monophyletic [4,5]. Equally uncontroversial is the evidence that many eukaryotic lineages have independently evolved some form of multicellularity [6–8]. To reflect the ease of this transition, Grosberg & Strathmann [7] characterize multicellularity as a ‘minor major transition’. Indeed, a recent study claims to have evolved multicellular yeast in the laboratory de novo [9]. Consequently, the study of the early evolution of multicellular organisms focuses on environmental ‘triggers’ [10,11].
The origins of both eukaryotes and multicellularity may have been driven by the advantages of larger size (efficient dispersal, exploitation of more or different food sources, producing more offspring, escaping predators, avoidance of the constraints of low Reynolds numbers) [6,12,13]. Both transitions were initiated by clever engineering solutions to surface-to-volume constraints. In the case of eukaryotes, energy-converting membranes were moved internally [14]. In the case of multicellularity, other surface- and size-dependent processes were facilitated [12].
The origins of both eukaryotes and multicellularity bolster the conceptualization of the history of life as repeated transitions between levels or units of selection [15–22]. In the former, bacterial cells formed the collective; in the latter, eukaryotic cells formed the collective. The multi-level theory of evolution provides a framework for understanding these transitions. Central to this understanding are sequential stages of cooperation, conflict and conflict mediation [23]. Cooperation among lower-level units leads to nascent higher-level units. Conflicts among lower-level units can then undermine these emerging units. If conflicts can be mediated, however, then fully formed higher-level units can emerge. These new units can outcompete individual lower-level units and proliferate. Subsequently, if conflicts can again be mediated, these higher-level units themselves can band together to form even higher-level units. The history of life can thus be viewed as a repetition of stages of cooperation leading to conflict, and conflict mediation leading to emergence [23]. This process underlies many transitions in evolution, and these transitions are thus in some sense equivalent [16,22].
Or are they equivalent? Biological details can complicate these theoretical generalities [22]. So the question remains—if the evolutionary issues were similar (i.e. reconciling conflict among lower-level units in favour of the higher-level unit), why did the eukaryotic cell evolve only once, while by contrast multicellular eukaryotes have evolved many times? A general theoretical framework may only provide limited insights into a particular transition. Key biological details—particularly the nature of the conflicts and the available mechanisms for conflict mediation—need to be elucidated. In the case of eukaryotes, there may have been something achieved in their origin that facilitates the subsequent origin of multicellularity. Re-examining the nature of the eukaryotic cell in relation to conflict and conflict mediation can provide insights into what this something is.
2. Conflict and conflict mediation and the endosymbiont theory
Early-twentieth century formulations of the endosymbiont theory of the origin of eukaryotes explicitly rejected ‘Darwinian’ notions of conflict and posed cooperation as an alternative. Such views were common particularly in Russia [24]. For example, Mereschkowsky [25] makes no mention of conflict (but see [26]). Later, Wallin [27] described ‘symbionticism’ as a missing part of Darwin's theory, seemingly outside the realm of natural selection. Margulis [28] also had little to say about potential conflicts, despite discussing scenarios in which conflict would seem inevitable. In her later writings, Margulis’ view of conflict remained unchanged, for example, Margulis and Sagan: ‘ … the view of evolution as chronic bloody competition…dissolves before a new view of continual cooperation … Life did not take over the globe by combat, but by networking’ [29, pp. 14–15; 30, p. 11].
Later theorists of course did raise the issue of conflict—both in general, and with regard to mitochondria, in particular. Cosmides & Tooby [31] rigorously examined the potential for intragenomic conflict within a eukaryotic cell. Williams [32, p. 42] notes ‘the subsequent stability of these eukaryotic cell lineages through geologic time, despite potential disruption from selection among cellular components, presents an evolutionary problem that deserves detailed attention’. From the perspective of mitochondrial physiology, Blackstone [33] addressed this question as well. Nevertheless, these theoretical considerations had relatively little impact. Rather, it was the empirical discovery of the role of mitochondria in programmed cell death [34,35] that firmly established the notion of conflictual relationships in the formation of the eukaryotic cell [36–38]. In the following, conflict and its mediation are further explored.
3. Conflict and conflict mediation and the origin of eukaryotes: a closer look
A proto-eukaryote can be conceptualized as a colony of proto-mitochondria within a larger cell that also contained the host genome. Superficially, this seems quite similar to standard levels-of-selection models [22] in which particles are nested within a collective (figure 1a). The host genome could be considered a unique particle, whereas the proto-mitochondria would comprise an interchangeable group of particles. Nevertheless, such a conceptualization leaves out a critical feature of the proto-eukaryote: the cytosol (figure 1b). Indeed, it is through the cytosol that the lower-level units or particles actually interact with each other. During metabolism, proto-mitochondria take up and emit various molecules; dying proto-mitochondria release DNA; the host genome takes up DNA and emits various gene products, and so on. To reconcile these considerations with levels-of-selection models, the cytosol could be regarded as an emergent feature or features [22]. This view of the cytosol is of particular significance to considerations of energetic regulation, as discussed in the following.
Figure 1.
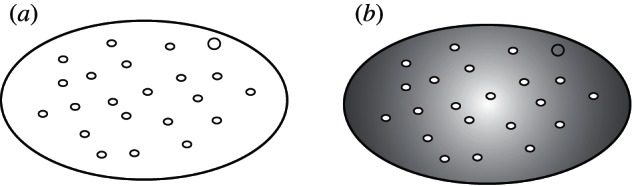
Levels-of-selection analyses usually focus on particles nested within a collective (a), for example, individuals within a society, or cells within an organism. In the case of proto-mitochondria within a proto-eukaryote, much of the collective is not contained within the particles, as emphasized in (b) with the cytosol shaded. The cytosol can be viewed as an emergent feature of the collective. The host genome appears as the unique particle.
Metabolic complementation is common in the microbial world [39]. In the initial association that led to eukaryotes, host and symbiont could have interacted in a variety of ways [3]. Nevertheless, there was one key requirement: in order for the eukaryote to attain larger size, the process of energy conversion had to be shifted to internal membranes [14]. Moving energy-converting symbionts inside a larger host is one way to accomplish this. In this scenario, however, the lower-level units now carry out energy conversion and allocation. Because replication requires energy, selection on the proto-mitochondria would inexorably favour allocation of energy into selfish replication. This is the principal conflict in the origin of eukaryotes. Genomic and energetic factors both have the potential to mediate this conflict.
Before considering these factors, a caveat should be noted. The diversity of modern mitochondria is only beginning to be characterized (e.g. genome size and structure, inheritance, recombination, signalling and so on [40–42]). Such information coupled with a well-resolved phylogeny of the eukaryotes could, in principle, be used to reconstruct the character states of the last common ancestor of modern mitochondria. Such an ancestor would still be highly derived compared with the first proto-mitochondrion [1], and modern out-groups are highly derived as well (figure 2). Hence, the following discussion is based on generalizations that seem supportable at this time [43].
Figure 2.
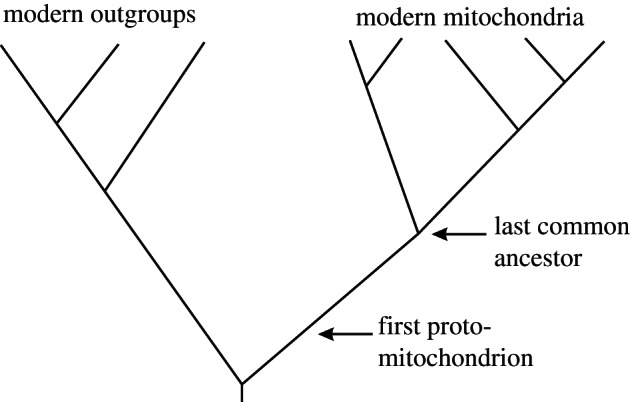
Evolutionary history of mitochondria. If the character states of all modern mitochondria were known, then features of the last common ancestor could be accurately reconstructed. Nevertheless, the last common ancestor may be very distinct from the first proto-mitochondrion (i.e. extensive evolution occurred in the mitochondrial stem group). Modern out-groups are also highly derived.
(a). Genetic and genomic factors
Restraining the selfish replication of proto-mitochondria has long been attributed to the formation of a chimeric (i.e. host/symbiont) nuclear genome, and the consequent reduction in the symbiont's genome and hence mutational space [13]. Nevertheless, organelles with functioning electron transport chains always retain a small genome [44,45], so the potential for heritable variation still exists even in modern mitochondria. Modern mitochondrial genomes generally seem to lack key factors involved in replication and energy allocation. Variant mitochondria that initiate or freely allocate energy into their own replication thus cannot evolve. Consequently, genome transfer does seem to have a role in mediating conflicts in modern mitochondria.
Such features of modern mitochondria notwithstanding, it remains difficult to see how genome transfer (at least by itself) could have regulated the conflicts during the origin of the eukaryotes. Genome transfer requires a series of evolutionary innovations, and none of these existed before the endosymbiosis. Prior to the evolution of the nucleus [46], it was likely a simple matter for bulk DNA released by damaged proto-mitochondria to incorporate into the host genome. Indeed, in this way, the host genome quickly gave rise to the chimeric, proto-nuclear genome. Recombination could activate the newly included genes by association with promoters. Symbiont genes thus transferred could then be expressed in the cytosol [47,48]. Some advantages might accrue to the higher-level unit by this transfer of symbiont biochemical pathways. Nevertheless, the effects of such transfer were likely minimal in terms of mediating conflicts: subsequent to the transfer, viable symbionts still retained a full complement of genes and a broad mutational space for selfish variants to arise.
To diminish this mutational space, it was necessary not only to transfer symbiont genes to the chimeric, proto-nuclear genome, but also to have their products re-enter the proto-mitochondria. This requires the evolution of the protein-import apparatus, and the association of proto-mitochondrial genes in the chimeric genome with appropriate transit peptides. If the gene product could be re-imported into a proto-mitochondrion, then the organelle copy could then become defective via mutation and ultimately lost. This will only occur, however, if there is no selection on the proto-mitochondrion to maintain a functional copy. In other words, the key question is whether a symbiont with a silenced gene loses out in competition with one in which this gene remains intact.
Consider, for instance, replication factors in this context. In a cell in which a gene for a replication factor has been transferred to the host genome and a re-import mechanism evolved, proto-mitochondria can lose this gene and still successfully replicate. Their replication can now be regulated by the higher-level unit. On the other hand, a proto-mitochondrion that retains the gene and selfishly replicates will increase in frequency and come to predominate within the collective. It is thus difficult to see how the system of gene transfer could be used to modulate the selfish replication of the lower-level units. Indeed, the argument seems to be perfectly circular: relaxed selection is necessary for gene loss, and gene loss is necessary for relaxed selection. Perhaps selection at the level of the mitochondrial genome or random processes, or both, can be invoked, leading to the fixation of proto-mitochondria missing the gene in a particular proto-eukaryotic cell [49,50]. The consequent advantages of that cell vis-à-vis its competitors could then favour its proliferation. Selection on the higher-level unit favouring genome transfer thus must be invoked to overcome selection on the lower-level units opposing such transfer. Nevertheless, in the early stages of an evolutionary transition, selection on the higher-level unit is likely to be weak relative to selection on the lower-level units [23]. In comparable circumstances, naïve ‘for the good of the group’ evolutionary scenarios have been heavily criticized previously [51,52].
In this context, functional advantages of the shift of most of the proto-mitochondrial genome to the nucleus may be relevant. A compact mitochondrial genome eliminates energetically expensive redundancy throughout the cell [1,2]. While in modern eukaryotes the loss of much of the mitochondrial genome may currently serve to mediate conflicts, this loss may not have evolved in this context. Rather, relocation of the genome may have evolved for functional benefits and may now mediate conflicts as a by-product.
(b). Energetic and metabolic factors
Because the functions of living cells require energy, metabolic regulation can be expected to have a central role in modern cells, and indeed it does [53,54]. Such metabolic regulation is likely a shared primitive feature of all cells [55]. In particular, because both growth and replication require energy, both are expected to be regulated by metabolic state [1]. Metabolic state, in turn, depends on a series of redox couples whose sources and sinks are environmental [44]. Thus, the growth and replication of all cells are ultimately regulated by the environment. In the case of proto-mitochondria, subsequent to the endosymbiosis, their environment was the cytosol of the proto-eukaryote. The content of the cytosol in terms of electron donors and acceptors would regulate the capacity of proto-mitochondria for growth and replication. For instance, an abundance of substrate would facilitate their replication, whereas a shortage of substrate would have an inhibitory effect.
As pointed out earlier, the cytosol was an emergent feature of the collective. The ingestions and excretions of the proto-mitochondria (and the chimeric, proto-nuclear genome) would influence its contents. If the scale of the cytoplasm was vastly greater than any proto-mitochondrion, however, no single lower-level unit could unduly control its contents. Thus, the proto-mitochondrial collective produces the metabolic state of the proto-eukaryote, and this metabolic state, in turn, regulates the replication of the members of the collective. Such metabolic regulation would be in place from the first day of the endosymbiosis—nothing had to be invented. During initial associations of host and symbiont, crucial conflict mediation can thus occur. Subsequently, as the higher-level unit began to emerge, metabolic regulation may continue to mediate conflict. The following scenarios illustrate these principles for two sequential stages very early in the evolution of eukaryotes.
(c). Stage 1: first steps
Endosymbioses in prokaryotes occur rarely [1]. Consider the first proto-mitochondrion, which by whatever means became endosymbiotic (figure 3). Some kind of metabolic, mutualistic relationship is assumed, but remains unspecified. For instance, the proto-mitochondrion may take up the reduced form of an electron carrier, oxidize it, and then excrete the oxidized form as waste, whereas in the cytosol, this molecule serves as an electron acceptor. In such a system, stoichiometry mediates conflict. In order to obtain energy, a proto-mitochondrion needs to oxidize the substrate, and hoarding the waste product would have negative fitness consequences. A proto-mitochondrion could evolve to oxidize substrate faster, but it would continue to emit waste in proportion to the amount of substrate oxidized. This would be adaptation, not defection. Greater efficiency of both partners would quickly evolve.
Figure 3.
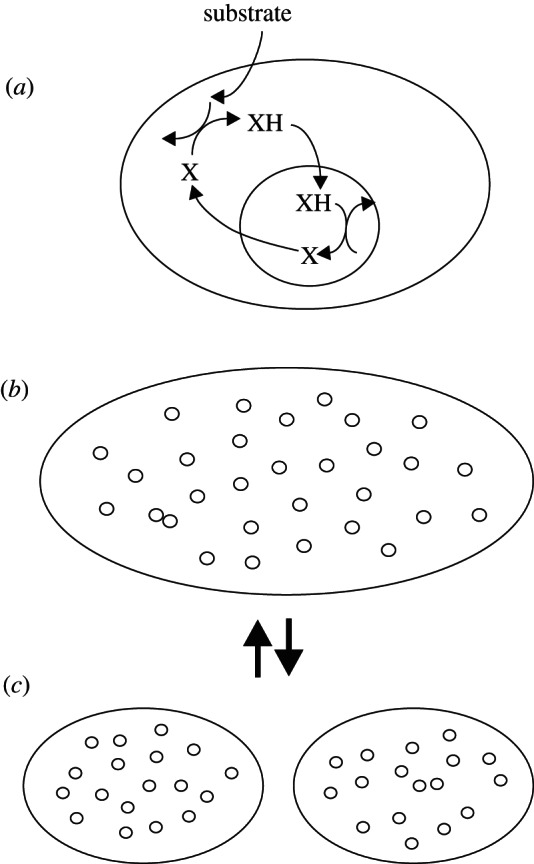
Stoichiometric mediation of conflict. In the initial endosymbiosis (a), the proto-eukaryote takes up substrate and uses an unspecified molecule, X, as a terminal electron acceptor. The proto-mitochondrion takes up the reduced form of this electron carrier, XH, and oxidizes it, excreting X. Both partners obtain energy in this fashion. The proto-mitochondrion replicates and the host increases in size (b), eventually dividing (c). This simple life cycle continues with conflicts mediated by stoichiometry.
Given such metabolic complementation, the initial proto-mitochondrion would find itself in a relatively rich environment. Rapid replication would ensue. Meanwhile, if the proto-eukaryote was provided with abundant substrate, then the ready availability of electron acceptors would also be advantageous. Indeed, if the proto-eukaryote did not use its external membrane in energy conversion, then size increase in proportion to the proto-mitochondrial population may also occur. Size increase of the proto-eukaryote could then provide new food resources [6,12]. At some point, the proto-eukaryote would attain a size where other surface-dependent processes became limiting. Proto-eukaryote division and assortment of proto-mitochondria would restore favourable conditions. With the attainment of this simple life cycle, the population of proto-eukaryotes would grow. The resulting large population would increase the probability of subsequent emergence.
(d). Stage 2: adenine nucleotide translocator and conflict mediation
As the proto-eukaryote evolved, no innovation was as seemingly fraught with peril for the collective as the adenine nucleotide translocator, ANT. ANT actively exports the metabolic currency of cells, ATP, from mitochondria to the cytosol, while taking up ADP. The evolution of ANT completely recasts the relationship between the proto-mitochondria and the proto-eukaryote. No longer are the former taking up something useful (e.g. XH) and excreting something useless (X), as in the case of simple metabolic complementation (figure 3). On the other hand, the major benefit of the symbiosis (specialization of energy conversion on internal membranes) cannot be realized without ANT. A benefit for the lower-level units can be realized as well, if proto-mitochondria are digested under stressful conditions in a process analogous to mitophagy [56], i.e. proto-mitochondria can emit ATP to stabilize the host and avoid digestion. As discussed earlier with genetic factors, however, these advantages accrue to the higher-level unit. Emitting ATP would seem to be vulnerable to defectors in a frequency-dependent fashion, i.e. a proto-mitochondrion with a disabled ANT will be at a selective advantage.
Or will it? As developed in scenario 1, increased efficiency follows from stoichiometric conflict mediation. One result would be ‘substrate shovelling’ from the plasma membrane to the proto-mitochondria. In these circumstances, ATP is abundantly produced, and a shortage of ATP is unlikely. Rather, the principal threat to the proto-mitochondrion is a shortage of metabolic demand. In this context, ANT may have evolved as a metabolic ‘safety valve’, creating reliable metabolic demand for the proto-mitochondrion. Without ANT, a variant proto-mitochondrion could potentially convert all of its ADP to ATP. Membrane potential would then become maximal, electron carriers would become highly reduced, and reactive oxygen formation would be maximal (i.e. state 4 metabolism). Damage to the proto-mitochondrion would ensue. In a process analogous to modern mitophagy [57,58], selective digestion of the damaged proto-mitochondrion may then occur. By this view, ANT conveys an advantage to the individual proto-mitochondrion by sustaining a state 3 metabolism (e.g. high ADP/ATP and NAD+/NADH ratios, low to moderate reactive oxygen species).
Notably, in cells of modern animals, the master regulator of mitochondrial biogenesis, PGC-1α, triggers the formation of new mitochondria under conditions of state 3 metabolism [59,60]. Replication of state 3 proto-mitochondria may also have occurred. Under these conditions, a proto-mitochondrion without ANT, even if it somehow avoided damage and selective digestion, still could at best only match the replication rate of the cooperator proto-mitochondria. Thus, when the metabolic demand of the proto-eukaryote was high, both variant and normal proto-mitochondria grow and divide at maximal rates (figure 4a). Under other metabolic conditions (e.g. low amounts of substrate or of electron acceptors), none of the proto-mitochondria would grow and divide. The variant proto-mitochondrion and its descendants, however, would likely have an advantage when the metabolic demand of the proto-eukaryote was low. Such a proto-eukaryote might lose out in competition with more rapidly dividing proto-eukaryotes (i.e. selection on the higher-level unit again must be invoked). Alternatively, because the bulk of the proto-mitochondria would enter state 4, initiation of the sexual phase of the life cycle should follow [61] and may by chance produce a daughter proto-eukaryote without the energetically selfish proto-mitochondria (figure 4b; see also Szathmáry & Demeter [62]).
Figure 4.
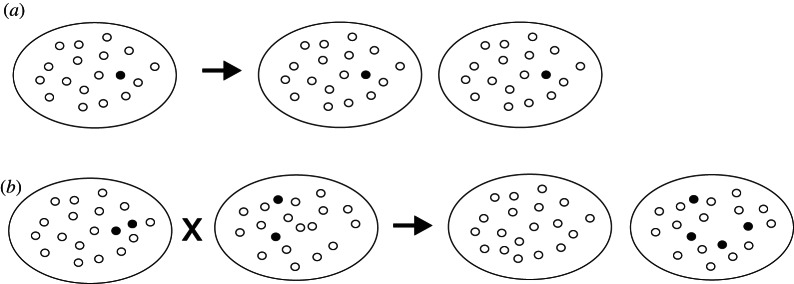
Regulation of energetically selfish proto-mitochondria by metabolic demand and stochastic processes. If the proto-eukaryote exerts strong metabolic demand because of rapid growth and replication (a), then normal proto-mitochondria (unfilled circles) will perceive a high ADP/ATP ratio and their rates of growth and division will match those of variant proto-mitochondria (filled circles). If metabolic demand of the proto-eukaryote falters and it undergoes whole-cell fusion and whole-genome recombination (b), stochastic processes may still produce a daughter cell with all normal proto-mitochondria.
Clearly, the evolution of ANT subsequently required mechanisms of conflict mediation more complex than mere stoichiometry. A way forward for the proto-eukaryote, however, can be envisioned by linking features of the life cycle of the collective to metabolic state:
state 3 metabolism of the collective—replication of the proto-eukaryote and proto-mitochondria,
state 4 metabolism of a lower-level unit—mitophagy of an individual proto-mitochondrion and
state 4 metabolism of the collective—sex involving whole-cell fusion of two proto-eukaryotes and stochastic reapportionment of proto-mitochondria to the daughter cells (figure 4b).
These generalities are supported by some [57–60,63] but not all [64,65] of the available data. Further investigations, particularly of early-diverging eukaryotes, are clearly necessary.
4. Eukaryotes and multicellular organisms: parallels and differences
Moving energy-converting membranes internally allowed eukaryotes to transcend surface-to-volume constraints. This clever engineering solution, however, turned into a levels-of-selection nightmare: the lower-level units could selfishly allocate energy into their own replication. This was the principal conflict in the origin of eukaryotes. Genomic transfer was likely not initially effective in mediating this conflict. On the other hand, the metabolic regulation inherent in the electron transport chain itself may have allowed sufficient conflict mediation. Nevertheless, the evolutionary requirements are stringent and many evolutionary experiments in forming complex cells may have failed to mediate these conflicts and ended in failure [1].
In the transition to multicellularity, lower-level units again can take up more than their share of substrate and allocate it into selfish replication. Because there is no structure analogous to the nucleus, genome transfer is impossible, and genomic deletion in somatic cells is not commonly found. Metabolic regulation, however, may be achieved by extending within-cell signalling derived during the conflictual stages of the origin of eukaryotes to between-cell signalling. The extent to which mitochondrial pathways connect to within- and between-cell signalling may provide an indication that this has occurred (table 1). These may be the features of ‘genomic and bioenergetic complexity’ derived by eukaryotes that allowed complex multicellularity to evolve repeatedly [2].
Table 1.
A list of putative exemplars of signalling molecules that are both connected to mitochondrial metabolism and also used in between-cell signalling.
| molecule | mitochondrial function | other functions |
|---|---|---|
| cAMP | modulates electron transport chain [66] | second messenger [66] |
| ATP | signals metabolic state (see text) | purinergic signalling [67] |
| reactive oxygen species | signals metabolic state (see text) | disulfide relays [68] |
| calcium ions | metabolic activator [69] | second messenger [70] |
| STAT3 | component of electron transport chain [71] | JAK-STAT pathway [72] |
| cytochrome c | component of electron transport chain [73] | apoptosis [34,35] |
| VEGF | regulator of fatty acid metabolism [74] | angiogenesis [75] |
| insulin | activator of pyruvate dehydrogenase complex [76] | hormone regulating carbohydrate and fat metabolism [77] |
| p53 | metabolic regulation [78] | tumour suppressor [79] |
In conclusion, any evolutionary transition in which the lower-level units carry out energy conversion and allocation will be extraordinarily challenging. This is the central reason why eukaryotes only evolved once. Yet by successfully mediating these conflicts, eukaryotes may have paved the way for repeated evolution of multicellularity, which could occur simply by coopting the existing within-cell mechanisms of conflict mediation into between-cell ones.
Acknowledgements
Many thanks to the organizers of the discussion and satellite meetings. Other participants and two reviewers provided helpful comments, and the NSF provided support (grant no. EF-0531654).
References
- 1.Lane N. 2011. Energetics and genetics across the prokaryote–eukaryote divide. Biol. Direct. 6, 35. 10.1186/1745-6150-6-35 (doi:10.1186/1745-6150-6-35) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane N, Martin W. 2010. The energetics of genome complexity. Nature 467, 929–934 10.1038/nature09486 (doi:10.1038/nature09486) [DOI] [PubMed] [Google Scholar]
- 3.Embley TM, Martin W. 2006. Eukaryotic evolution, changes and challenges. Nature 440, 623–630 10.1038/nature04546 (doi:10.1038/nature04546) [DOI] [PubMed] [Google Scholar]
- 4.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290, 972–977 10.1126/science.290.5493.972 (doi:10.1126/science.290.5493.972) [DOI] [PubMed] [Google Scholar]
- 5.Katz LA, Grant JR, Parfrey LW, Burleigh JG. 2012. Turning the crown upside down: gene tree parsimony roots the eukaryotic tree of life. Syst. Biol. 61, 653–660 10.1093/sysbio/sys026 (doi:10.1093/sysbio/sys026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonner JT. 1998. The origins of multicellularity. Integr. Biol. 1, 27–36 (doi:10.1002/(SICI)1520-6602(1998)1:1<27::AID-INBI4>3.0.CO;2-6) [DOI] [Google Scholar]
- 7.Grosberg RK, Strathmann RR. 2007. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Syst. 38, 621–654 10.1146/annurev.ecolsys.36.102403.114735 (doi:10.1146/annurev.ecolsys.36.102403.114735) [DOI] [Google Scholar]
- 8.Rokas A. 2008. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu. Rev. Genet. 42, 235–251 10.1146/annurev.genet.42.110807.091513 (doi:10.1146/annurev.genet.42.110807.091513) [DOI] [PubMed] [Google Scholar]
- 9.Ratcliff WC, Denison RF, Borrello M, Travisano M. 2012. Experimental evolution of multicellularity. Proc. Natl Acad. Sci. USA 109, 1595–1600 10.1073/pnas.1115323109 (doi:10.1073/pnas.1115323109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canfield DE, Poulton SW, Narbonne GM. 2007. Late-Neoproterozoic deep-ocean oxygenation and the rise of animal life. Science 315, 92–95 10.1126/science.1135013 (doi:10.1126/science.1135013) [DOI] [PubMed] [Google Scholar]
- 11.Boyle RA, Lenton TM, Williams TP. 2007. Neoproterozoic ‘snowball Earth’ glaciations and the evolution of altruism. Geobiology 5, 337–349 10.1111/j.1472-4669.2007.00115.x (doi:10.1111/j.1472-4669.2007.00115.x) [DOI] [Google Scholar]
- 12.Bonner JT. 1988. The evolution of complexity. Princeton, NJ: Princeton University Press [Google Scholar]
- 13.Lachmann M, Blackstone NW, Haig D, Kowald A, Michod RE, Szathmáry E, Werren JH, Wolpert L. 2003. Group report, Cooperation and conflict in the evolution of genomes, cells, and multicellular organisms. In Genetic and cultural evolution of cooperation (ed. Hammerstein P.), pp. 327–356 Cambridge, MA: MIT Press [Google Scholar]
- 14.Lane N. 2005. Power, sex, suicide: mitochondria and the meaning of life. Oxford, UK: Oxford University Press [Google Scholar]
- 15.Buss L. 1987. The evolution of individuality. Princeton, NJ: Princeton University Press [Google Scholar]
- 16.Maynard Smith J, Szathmáry E. 1995. The major transitions in evolution. Oxford, UK: Oxford University Press [Google Scholar]
- 17.Maynard Smith J, Szathmáry E. 1999. The origins of life. Oxford, UK: Oxford University Press [Google Scholar]
- 18.Keller L. (ed). 1999. Levels of selection in evolution. Princeton, NJ: Princeton University Press [Google Scholar]
- 19.Michod RE. 1999. Darwinian dynamics: evolutionary transitions in fitness and individuality. Princeton, NJ: Princeton University Press [Google Scholar]
- 20.Queller DC. 2000. Relatedness and the fraternal major transitions. Phil. Trans. R. Soc. Lond. B 355, 1647–1655 10.1098/rstb.2000.0727 (doi:10.1098/rstb.2000.0727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerstein P. 2003. Genetic and cultural evolution of cooperation. Cambridge, MA: The MIT Press [Google Scholar]
- 22.Okasha S. 2006. Evolution and the levels of selection. Oxford, UK: Oxford University Press [Google Scholar]
- 23.Michod RE, Nedelcu AM. 2003. On the reorganization of fitness during evolutionary transitions in individuality. Integr. Comput. Biol. 43, 64–73 10.1093/icb/43.1.64 (doi:10.1093/icb/43.1.64) [DOI] [PubMed] [Google Scholar]
- 24.Dugatkin LA. 2011. The prince of evolution. Seattle, WA: CreateSpace [Google Scholar]
- 25.Mereschkowsky C. 1905. Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol. Centralbl. 25, 593–604 (English translation in Martin W and Kowallik KV. 1999 Eur. J. Phycol.34, 287–295.) [Google Scholar]
- 26.Sapp J, Carrapiço F, Zolotonosov M. 2002. Symbiogenesis: the hidden face of Constantin Merezhkowsky. Hist. Phil. Life Sci. 24, 413–440 10.1080/03919710210001714493 (doi:10.1080/03919710210001714493) [DOI] [PubMed] [Google Scholar]
- 27.Wallin IE. 1927. Symbionticism and the origin of species. Baltimore, MD: Williams & Wilkins [Google Scholar]
- 28.Margulis L. 1970. Origin of eukaryotic cells. New Haven, CT: Yale University Press [Google Scholar]
- 29.Margulis L, Sagan D. 1986. Microcosmos. New York, NY: Summit [Google Scholar]
- 30.Margulis L, Sagan D. 2001. Marvellous microbes. Resurgence 206, 10–12 [Google Scholar]
- 31.Cosmides LM, Tooby J. 1981. Cytoplasmic inheritance and intragenomic conflict. J. Theor. Biol. 89, 83–129 10.1016/0022-5193(81)90181-8 (doi:10.1016/0022-5193(81)90181-8) [DOI] [PubMed] [Google Scholar]
- 32.Williams GC. 1992. Natural selection. Oxford, UK: Oxford University Press [Google Scholar]
- 33.Blackstone NW. 1995. A units-of-evolution perspective on the endosymbiont theory of the origin of the mitochondrion. Evolution 49, 785–796 10.2307/2410402 (doi:10.2307/2410402) [DOI] [PubMed] [Google Scholar]
- 34.Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. 1997. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science 275, 1132–1136 10.1126/science.275.5303.1132 (doi:10.1126/science.275.5303.1132) [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Liu X, Bhalla K, Kim CN, Ibrado AM, Cai J, Peng TI, Jones DP, Wang X. 1997. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 275, 1129–1132 10.1126/science.275.5303.1129 (doi:10.1126/science.275.5303.1129) [DOI] [PubMed] [Google Scholar]
- 36.Frade JM, Michaelidis TM. 1997. Origin of eukaryotic programmed cell death: a consequence of aerobic metabolism. BioEssays 19, 827–832 10.1002/bies.950190913 (doi:10.1002/bies.950190913) [DOI] [PubMed] [Google Scholar]
- 37.Kroemer G. 1997. Mitochondrial implication in apoptosis: towards an endosymbiont hypothesis of apoptosis evolution. Cell Death Differ. 4, 443–456 10.1038/sj.cdd.4400266 (doi:10.1038/sj.cdd.4400266) [DOI] [PubMed] [Google Scholar]
- 38.Mignotte B, Vayssiere J-L. 1998. Mitochondria and apoptosis. Eur. J. Biochem. 252, 1–15 10.1046/j.1432-1327.1998.2520001.x (doi:10.1046/j.1432-1327.1998.2520001.x) [DOI] [PubMed] [Google Scholar]
- 39.Fenchel T, Finlay BJ. 1995. Ecology and evolution in anoxic worlds. Oxford, UK: Oxford University Press [Google Scholar]
- 40.Lang BF, Burger G, O'Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW. 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387, 493–497 10.1038/387493a0 (doi:10.1038/387493a0) [DOI] [PubMed] [Google Scholar]
- 41.Barr CM, Neiman M, Taylor DR. 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi, and animals. New Phytol. 168, 39–50 10.1111/j.1469-8137.2005.01492.x (doi:10.1111/j.1469-8137.2005.01492.x) [DOI] [PubMed] [Google Scholar]
- 42.Collins SR, Meyer T. 2011. Evolutionary origins of STIM1 and STIM2 within ancient Ca2+ signaling systems. Trends Cell Biol. 21, 202–211 10.1016/j.tcb.2011.01.002 (doi:10.1016/j.tcb.2011.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller M, et al. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495 10.1128/MMBR.05024-11 (doi:10.1128/MMBR.05024-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen JF. 1993. Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J. Theor. Biol. 165, 609–631 10.1006/jtbi.1993.1210 (doi:10.1006/jtbi.1993.1210) [DOI] [PubMed] [Google Scholar]
- 45.Allen JF. 2003. The function of genomes in bioenergetic organelles. Phil. Trans. R Soc. Lond. B 358, 19–38 10.1098/rstb.2002.1191 (doi:10.1098/rstb.2002.1191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin W, Koonin EV. 2006. Introns and the origin of nucleus–cytosol compartmentalization. Nature 440, 41–45 10.1038/nature04531 (doi:10.1038/nature04531) [DOI] [PubMed] [Google Scholar]
- 47.Martin W, Herrmann RG. 1998. Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 118, 9–17 10.1104/pp.118.1.9 (doi:10.1104/pp.118.1.9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henze K, Martin W. 2001. How do mitochondrial genes get into the nucleus? Trends Genet. 17, 383–387 10.1016/S0168-9525(01)02312-5 (doi:10.1016/S0168-9525(01)02312-5) [DOI] [PubMed] [Google Scholar]
- 49.Rand DM. 2001. The units of selection on mitochondrial DNA. Annu. Rev. Ecol. Syst. 32, 415–448 10.1146/annurev.ecolsys.32.081501.114109 (doi:10.1146/annurev.ecolsys.32.081501.114109) [DOI] [Google Scholar]
- 50.Kowald A, Kirkwood TBL. 2011. Evolution of the mitochondrial fusion–fission cycle and its role in aging. Proc. Natl Acad. Sci. USA 108, 10 237–10 242 10.1073/pnas.1101604108 (doi:10.1073/pnas.1101604108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maynard Smith J. 1964. Group selection and kin selection. Nature 201, 1145–1147 10.1038/2011145a0 (doi:10.1038/2011145a0) [DOI] [Google Scholar]
- 52.Williams GC. 1966. Adaptation and natural selection. Princeton, NJ: Princeton University Press [Google Scholar]
- 53.Lazar MA, Birnbaum MJ. 2012. De-meaning of metabolism. Science 336, 1651–1652 10.1126/science.1221834 (doi:10.1126/science.1221834) [DOI] [PubMed] [Google Scholar]
- 54.Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, Sweedler JV, Cox CL, Gilietter MU. 2012. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science 337, 839–842 10.1126/science.1222826 (doi:10.1126/science.1222826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blackstone NW. 2007. A food's-eye view of the transition from basal metazoans to bilaterians. Integr. Comput. Biol. 47, 724–733 10.1093/icb/icm056 (doi:10.1093/icb/icm056) [DOI] [PubMed] [Google Scholar]
- 56.Goldman SJ, Taylor R, Zhang Y, Jin S. 2010. Autophagy and the degradation to mitochondria. Mitochondrion 10, 309–315 10.1016/j.mito.2010.01.005 (doi:10.1016/j.mito.2010.01.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim I, Lemasters JJ. 2011. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antiox. Redox Signal. 14, 1919–1928 10.1089/ars.2010.3768 (doi:10.1089/ars.2010.3768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Twig G, Shirihai OS. 2011. The interplay between mitochondrial dynamics and mitophagy. Antiox. Redox Signal. 14, 1939–1951 10.1089/ars.2010.3779 (doi:10.1089/ars.2010.3779) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu Z, et al. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 10.1016/S0092-8674(00)80611-X (doi:10.1016/S0092-8674(00)80611-X) [DOI] [PubMed] [Google Scholar]
- 60.Arany Z, et al. 2008. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1α. Nature 451, 1008–1012 10.1038/nature06613 (doi:10.1038/nature06613) [DOI] [PubMed] [Google Scholar]
- 61.Blackstone NW, Kirkwood TBL. 2003. Mitochondria and programmed cell death: ‘slave revolt’ or community homeostasis? In Genetic and cultural evolution of cooperation (ed. Hammerstein P.), pp. 309–325 Cambridge, MA: MIT Press [Google Scholar]
- 62.Szathmáry E, Demeter L. 1987. Group selection of early replicators and the origin of life. J. Theor. Biol. 128, 463–486 10.1016/S0022-5193(87)80191-1 (doi:10.1016/S0022-5193(87)80191-1) [DOI] [PubMed] [Google Scholar]
- 63.Lin J, Handschin C, Spiegelman BM. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1, 361–370 10.1016/j.cmet.2005.05.004 (doi:10.1016/j.cmet.2005.05.004) [DOI] [PubMed] [Google Scholar]
- 64.Moreno-Loshuertos R, Acin-Pérez R, Fernández-Silva P, Movilla N, Pérez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enríquez JA. 2006. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 38, 1261–1268 10.1038/ng1897 (doi:10.1038/ng1897) [DOI] [PubMed] [Google Scholar]
- 65.Lane N. 2011. Mitonuclear match: optimizing fitness and fertility over generations drives ageing within generations. BioEssays 33, 860–869 10.1002/bies.201100051 (doi:10.1002/bies.201100051) [DOI] [PubMed] [Google Scholar]
- 66.Buck J, Levin LR. 2011. Physiological sensing of carbon dioxide/bicarbonate/pH via cyclic nucleotide signaling. Sensors 11, 2112–2128 10.3390/s110202112 (doi:10.3390/s110202112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng W, Watts LT, Holstein DM, Prajapati SI, Keller C, Grass EH, Walter CA, Lechleiter JD. 2010. Purinergic receptor stimulation reduces cytotoxic edema and brain infarcts in mouse induced by photothrombosis by energizing glial mitochondria. PLoS ONE 5, e14401. 10.1371/journal.pone.0014401 (doi:10.1371/journal.pone.0014401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Filomeni G, Rotilio G, Ciriolo MR. 2005. Disulfide relays and phosphorylation cascades: partners in redox-mediated signaling pathways. Cell Death Differ. 12, 1555–1563 10.1038/sj.cdd.4401754 (doi:10.1038/sj.cdd.4401754) [DOI] [PubMed] [Google Scholar]
- 69.Rizzuto R, et al. 2009. Ca2+ transfer from the ER to mitochondria: when, how, and why. Biochim. Biophys. Acta 1787, 1342–1351 10.1016/j.bbabio.2009.03.015 (doi:10.1016/j.bbabio.2009.03.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berridge MJ, Bootman MD, Roderick HL. 2003. Calcium signaling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529 10.1038/nrm1155 (doi:10.1038/nrm1155) [DOI] [PubMed] [Google Scholar]
- 71.Wegrzyn J, et al. 2009. Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797 10.1126/science.1164551 (doi:10.1126/science.1164551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levy DE, Lee C-k. 2002. What does Stat3 do? J. Clin. Invest. 109, 1143–1148 10.1172/JCI15650 (doi:10.1172/JCI15650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheffler IE. 1999. Mitochondria. New York, NY: John Wiley [Google Scholar]
- 74.Hagberg CE, et al. 2010. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature 464, 917–921 10.1038/nature08945 (doi:10.1038/nature08945) [DOI] [PubMed] [Google Scholar]
- 75.Tammela T, Enholm B, Alitalo K, Paavonen K. 2005. The biology of vascular endothelial growth factors. Cardiovasc. Res. 65, 550–563 10.1016/j.cardiores.2004.12.002 (doi:10.1016/j.cardiores.2004.12.002) [DOI] [PubMed] [Google Scholar]
- 76.Mukherjee C, Jungas RL. 1975. Activation of pyruvate dehydrogenase in adipose tissue by insulin. Biochem. J. 148, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simpson IA, Cushman SW. 1986. Hormonal regulation of mammalian glucose transport. Annu. Rev. Biochem. 55, 1059–1089 10.1146/annurev.bi.55.070186.005211 (doi:10.1146/annurev.bi.55.070186.005211) [DOI] [PubMed] [Google Scholar]
- 78.Khutornenko AA, Roudko VV, Chernyak BV, Vartapetian AB, Chumakov PM, Evstafieva AG. 2010. Pyrimidine biosynthesis links mitochondrial respiration to the p53 pathway. Proc. Natl Acad. Sci. USA 107, 12 828–12 833 10.1073/pnas.0910885107 (doi:10.1073/pnas.0910885107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. 2005. The antioxidant function of the p53 tumor suppressor. Nat. Med. 11, 1306–1313 10.1038/nm1320 (doi:10.1038/nm1320) [DOI] [PMC free article] [PubMed] [Google Scholar]


