Abstract
The physical and chemical mechanisms of small molecules with pharmacological activity forming nano-structures are developing into a new field of nano-medicine. By using ROESY 2D NMR spectroscopy, trandem mass spectroscopy, transmission electron microscopy and computer-assisted molecular modeling, this paper demonstrated the contribution of the folded conformation, the intra- and intermolecular π-π stacking, the intra- and intermolecular hydrogen bonds, and the receptor binding free energy of 6-dimethylaminonaph-2-yl-{N-S-[1-benzylcarba-moyl-4-(2-chloroacetamidobutyl)]-carboxamide (YW3-56) to the rapid formation of nano-rings and the slow formation of nano-capsules. Thus we have developed a strategy that makes it possible to elucidate the physical and chemical mechanisms of bioactive small molecules forming nano-structures.
Keywords: Mechanism, Nano-structure, Nano-medicine, Conformation, PAD4, Anti-cancer
1. INTRODUCTION
The nano-species of biological systems have been widely viewed in perspective.1-3 Various technologies have been used for preparing the nano-structures and assaying the properties of the nano-materials,4-11 which in turn promoted the developing of nano-medicine.12-15 In respect of the nano-medicine, in the past decades the development of the nano-technology mainly focused on the preparation of drug delivery systems, including preparing liposome.16,17 Recently the nano-scale assembly of single small molecule,18,19 and the effect of self-assembly of some bioactive small molecules on the promotion or the inhibition of their biological activities are under intense investigation.20,21 Of the achievements, a series of self-assembled bioactive small molecules have been explored to enhance the biological efficiency, such as in the functionalizing of metal surface,22 in the crafting functional biological materials,23 and in the increasing of the anti-osteoporosis activity.24 On the other hand, the physical and chemical mechanisms of the self-assembly of small molecules remain to be elucidated. This paper selected 6-dimethylaminonaph-2-yl-{N-S-[1-benzylcarbamoyl-4-(2-chloroacetamidobutyl)]-carboxamide (YW3-56),25 a small molecule capable of inhibiting cancer growth, as a model compound to explain the conformational basis of bioactive small molecules forming nano-structures and targeting the active site pocket of the enzyme peptidylarginine deiminase 4 (PAD4), and to elucidate the physical and chemical mechanisms of forming nano-structures.
2. EXPERIMENTAL SECTION
Synthesis
To conveniently obtain YW3-56 the 4-step route of the literature was followed.26 In step 1 and 2 the coupling reagent was changed from HATU and DIPEA to DCC and NMM. In step 1 and 2 the reaction solvent was changed from DMF to THF and from DMF to a mixed solvent of THF and DMF (1:1), respectively. Details of the synthetic route are provided in Supporting Information.
Conformation generation
YW3-56 was sketched in ChemDraw 10.0, converted to 3D conformation in Chem3D 10.0 and then energy minimized in Discovery Studio 2.1 with MMFF force field. The energy minimized conformation was utilized as the starting conformation for conformation generation of YW3-56. The energy-minimized conformations were sampled in the whole conformational space via systematic search method and BEST method in Discovery Studio 2.1. Both of the systematic search and BEST methods were performed with SMART minimizer using CHARMm force field. The energy threshold was set to 20 kcal/mol at 300 K. The maximum minimization steps were set to 200 and the minimization RMS gradient was set to 0.1 Å. The maximum generated conformations were set to 255 with a RMSD cutoff 0.2 Å.
1H NMR spectrum measurements
One-dimensional 1H NMR spectra were recorded on a Bruker 800 MHz spectrometer, ~5 mg of YW3-56 in 0.5 mL of deuteron dimethyl sulfoxide (DMSO-d6). The probe temperature was regulated to 298 K. The spectra were recorded by using a simple pulse-acquire sequence zg30. Typical acquisition parameters consisted of 64 K points covering a sweep width of 16447 Hz, a pulse width (pw90) of 8.63 μs and a total repetition time of 24 s to ensure full relaxation of the 1H resonances. Digital zero filling to 64 K and a 0.3 Hz exponential function were applied to the FID before Fourier transformation. The resonance at 2.5 ppm due to residual solvents, present as impurities (CD2HSOCD2H), tetramethylsilane (TMS) was used as internal reference. Standard absorptive two-dimensional 1H-1H chemical shift correlation spectra (COSY) were acquired with the same spectrometer. Each spectra consisted of a matrix of 2 K (F2) by 0.5 K (F1) covering a sweep width of 9615.4 Hz. Before Fourier transformation, the matrix was zero filled to 1 K by 1 K and standard sinebell apodization functions were applied in both dimensions. Two-dimensional ROESY experiments were acquired in the phase-sensitive mode using the same spectrometer. Each spectrum consisted of a matrix of 2 K (F2) by 1 K (F1) covering a sweep width of 9615.4 Hz. Spectra were obtained using spin-lock mixing periods of 200 ms. Before Fourier transformation, the matrix was zero filled to 1 K by 1 K and qsine apodization functions were applied in both dimensions. A full list of the 1HNMR, COSY and ROESY spectra are provided in Supporting Information.
Molecular docking towards PAD4
Software AutoDock 4 was used to perform the molecular docking of two folded conformations of YW3-56 towards the active site pocket of PAD4. It allows automated docking of flexible ligands to a rigid receptor with certain flexible residues. The receptor PAD4 was obtained from PDB entry 1WDA with the benzyol-L-Arg removed. The receptor PAD4 was treated as rigid and prepared by AutoDockTools 1.5: merging non-polar hydrogens, assigning gasteiger charges and autodock elements. The two folded conformations of YW3-56 were treated as flexible ligands and prepared by AutoDockTools 1.5: merging non-polar hydrogens, assigning gasteiger charges, finding root and aromatic carbons, detecting rotatable bonds and setting torsions. The grid box dimensions were set to 80 Å × 80 Å × 80 Å with a grid spacing of 0.375 Å. The grid box center was set to the center of benzyol-L-Arg in the crystal structure PDB 1WDA. The Lamarckian genetic algorithm (LGA) was used to find the appropriate binding positions, orientations, and the conformations of YW3-56 in the active site pocket of PAD4. The global optimization was started with parameters of a population of 300 randomly positioned individuals. The maximum number of energy evaluations was increased to 2.5×107 and the maximum number of generations in the LGA algorithm was increased to 2.7×105. The Solis & Wets local search was performed with a maximum number of 3,000. During the molecular docking experiments, 200 runs were carried out for each ligand. The resulted 200 conformations of each ligand were scored by the lowest binding energy and clustered with a rms tolerance of 2.0 Å.
Trandem mass spectroscopy test
Mass spectra were acquired using a solariX FT-ICR mass spectrometer (Bruker Daltonik) consisting of an ESI/MALDI dual ion source and 9.4 T superconductive magnet. Measurements were carried out in the positive MALDI ion mode. A Smart-beam-II laser (wavelength, 355 nm; focus setting, ‘medium’; repetition rate, 1000 Hz) was used as the ion source. The laser power was 40% and 1000 laser shots were accumulated for ions acquired. Q1 mass was set to 2467 m/z and isolation window was 5 m/z. Data were acquired by using solariXcontrol software. Spectral data were processed with Data Analysis software (Bruker Daltonik).
TEM measurements
The samples for transmission electron microscopy (TEM) measurements were prepared by dissolving YW3-56 in ethanol (analytical pure, 95%) to form a solution (0.1 μM). The solution was kept at room temperature for 30 min or was kept at 4 °C for one week. Then the solution was dropped onto the copper grids. The grids were then air-dried overnight. The nano-images were recorded by using TEM (Model JEM- 1230, JEOL).
AFM measurements
The samples for atomic force microscopy (AFM) measurements were prepared by dissolving YW3-56 in ethanol (analytical pure, 95%) to form a solution (0.1 μM). The solution was then dropped onto Mica discs and air-dried. The nano-images were recorded using the contact mode on a Nano-scope 3D AFM (Veeco Instruments Inc.).
Mean particle diameter measurements
The solution of YW3-56 (0.1 μM) were prepared in 95% of analytical pure ethanol at room temperature. After 30 min, the mean particle diameter was recorded every 30 s for 5 min on a Malvern’s Zeta Sizer (Nano-ZS90) with DTS (Nano) Program. The solution was kept at 4 °C for mean particle diameter measurement. Then the solution was measured for seven consecutive days according to the similar procedure.
SEM images of YW3-56 in solid state
The shape and size of the nano-species of the lyophilizing powders of 1 mM solutions of YW3-56 in ethanol were measured on a SEM (Hitachi S-4800, Hitachi High-Technologies Corp., Tokyo, Japan) at 2.0 kV. The lyophilizing powders were attached on to a copper plate via doubleside tape (Euromedex, France). The specimens were coated with 20 nm gold–palladium using a JEOL JFC-1600 AUTO FINE COATER. The coater was operated at 15 kV, 30 mA, 200 mTorr (argon) for 60 s. The shape and size distribution of the nanoparticles were measured from counting over 100 particles in randomly selected regions on the SEM alloy. All the measurements in triplicate grids were performed. The images were recorded on an imaging plate (Gatan Bioscan Camera Model1792) with 20 eV energy windows at 100 - 10 000 ×, and were digitally enlarged.
3. RESULTS AND DISCUSSION
3.1. Computer-assisted molecular modeling producing two folded conformations
To identify the conformation of YW3-56, computer-assisted molecular modeling was performed. The systematic search method produced 27 conformations, and the BEST method produced 183 conformations. The 210 generated conformations were visually examined and two conformations having the lowest free energy were selected. The conformation 1 has a −24.01 kcal/mol relative energy to the starting conformation (Figure 1a). The conformation 2 has a −16.4283 kcal/mol relative energy to the starting conformation (Figure 1b). In respect of the naphthyl and the benzyl, the two conformations produced via molecular modeling are folded ones.
Figure 1.
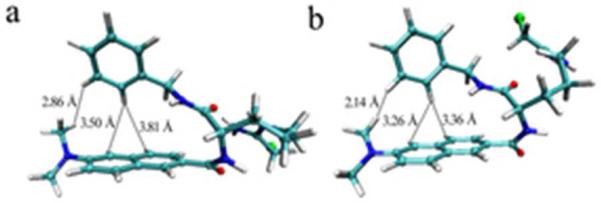
Two Low energy conformations 1 (a) and 2 (b) of YW3-56 from the systematic search method and the BEST method.
3.2. ROESY 2D NMR spectrum supporting folded conformation
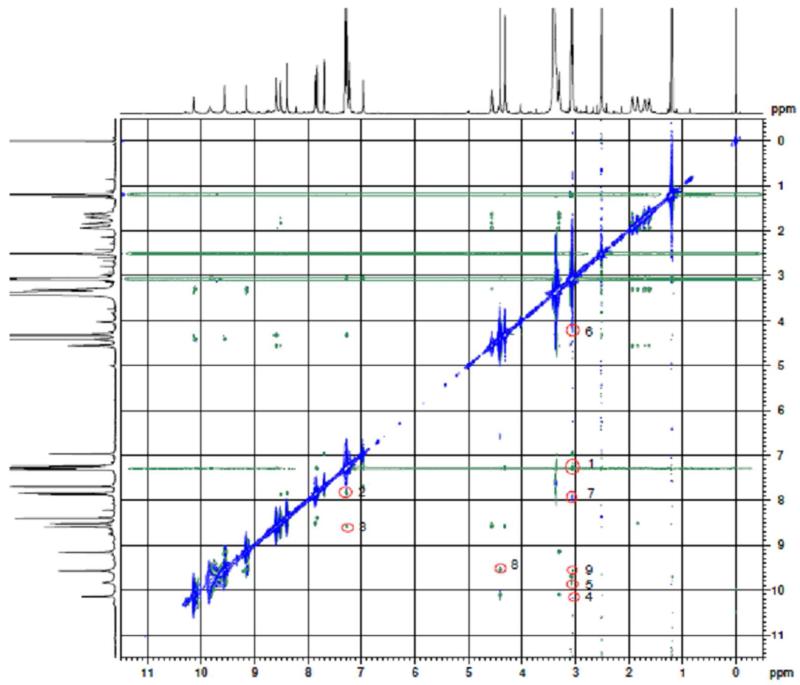
To confirm the folded conformations, the ROESY 2D NMR spectrum was measured and is shown in figure 2. In figure 2 nine cross-peaks were labeled with red circles. Of the nine cross-peaks, the cross-peaks 1-3 clearly support the folded conformations. The cross-peak 1 is for the H of the NCH3 and the H of the benzyl, the cross-peaks 2 and 3 are for the H of the naphthyl and the H of the benzyl. These cross-peaks indicate that the distance between the naphthyl and the benzyl is less than 4 Å. In this case the conformation must be a folded one. The π-π stacking should be the drive force for forming such a folded conformation.
Figure 2.
ROESY 2D NMR spectrum and the 9 cross-peaks. Cross-peaks 1-3 indicate that the intramolecular distance between the naphthyl and the benzyl is less than 4 Å. Cross-peaks 4-9 indicate the intermolecular interactions of YW3-56 molecules.
3.3. Molecular docking towards the active site pocket of PAD4 preferring folded conformation 2
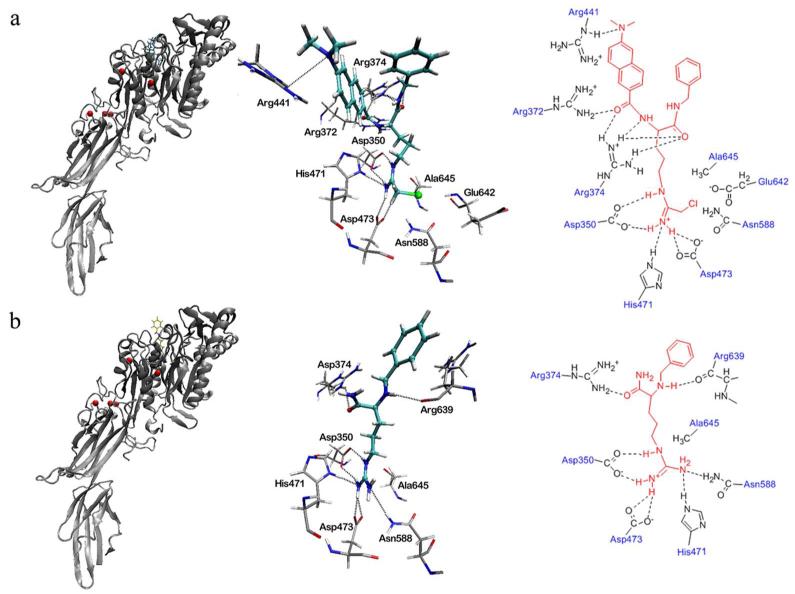
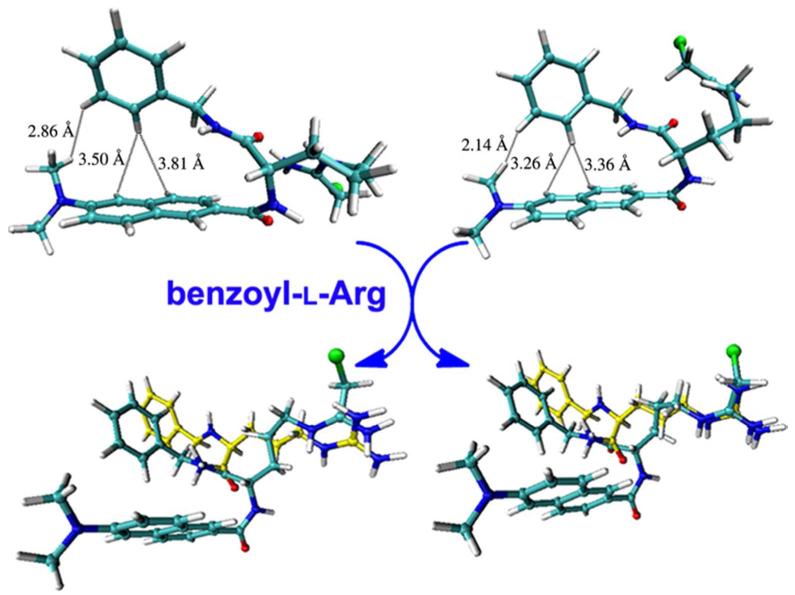
To pick a rational conformation from the two folded conformations, molecular docking of these two conformations towards the active site pocket of PAD4 was performed. This investigation gave conformations 1 and 2 the lowest binding free energy of −7.78 kcal/mol and −9.03 kcal/mol, respectively. This means that for the active site pocket of PAD4, conformation 2 is more suitable than 1. The binding mode of the folded conformation 2 in the active site pocket of PAD4 (Figure 3a) is similar to that of benzoyl-L-Arg in the crystal structure PDB 1WDA (Figure 3b). It was found that the modified arginine chain of YW3-56 inserts into the active site pocket of PAD4 and overlaps very well with benzoyl-L-Arg. The chloride of YW3-56 is close to residues Glu642, Asn588, and Asp473 with distances of 3.8, 3.12 and 4.02 Å. The modified arginine chain of YW3-56 and the arginine chain of benzoyl-L-Arg are positioned less than 4 Å from the active site residue Cys645 (Ala645 in Figure 3) and form hydrogen bonds with residues Asp350, His471, and Asp473. Besides, the 6-dimethylaminonaph-2-yl-carbonyl-amino moiety of YW3-56 has additional interactions such as the hydrogen bonds with residues Arg372, Arg374, Arg441 and Asp350of the pocket.
Figure 3.
The binding modes of the YW3-56 in the active site pocket of PAD4 (a) and benzoyl-L-Arg in the crystal structure of PDB 1WDA (b). The left panel of (a) and (b) show the binding site of YW3-56 (cyan bonds) and benzoyl-L-Arg (yellow bonds) at the active site pocket of PAD4 (gray ribbons); the middle panel of (a) and (b) show the 3D description of the hydrogen bond interactions between the ligands (cyan bonds) and PAD4 residues (gray bonds); the right panel of (a) and (b) show the planar description of the hydrogen bond interactions between ligands (red lines) and PAD4 residues (gray lines).
In further investigation, the binding mode of conformations 1 and 2 in the active site pocket of PAD4 were superimposed on benzoyl-L-Arg (yellow bonds in Figure 4), respectively. The modified arginine chain of the folded conformation 2 not the folded conformation 1 well overlapped to the arginine chain of benzoyl-L-Arg (Figure 4, right). Therefore the folded conformation 2 is considered more rational than conformation 1 to fit the active site pocket of PAD4.
Figure 4.
The superimposition of folded conformations 1 (left, cyan bonds) and 2 (right, cyan bonds) with benzoyl-L-Arg (yellow bonds) in the active site pocket of PAD4.
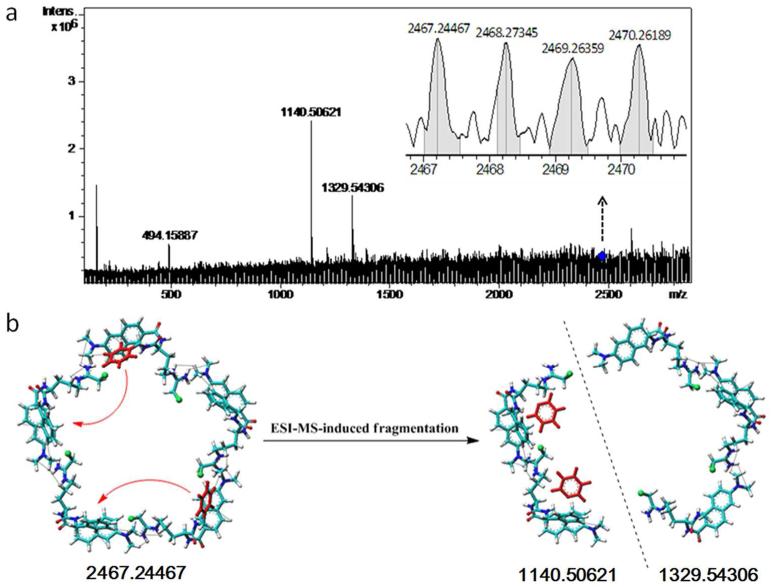
3.4. Trandem mass spectroscopy supporting YW3-56 existing as pentamer
In Figure 2 the cross-peaks 4 and 5 are for the H of the NCH3 of one molecule and the amide H of the chloroacetamidobutyl of another molecule, suggesting YW3-56 having intermolecular assembly. To know how many molecules were involved in this assembly, the trandem mass spectroscopy of Maldi with Quadrupole was tested. By using the daughter scanning model the ion (2466.24467 of mass-to-charge ratio) provided by the first scanning of Quadrupole-MS analyzer was isolated and collision splits. The fragment ions were scanned by the second FT-MS analyzer and two ions (1140.50621 and 1329.54306 of mass-to-charge ratio) were found (Figure 5a). There is a reasonable error (0.1954) between the [M]+ (2466.24467) and the two [M + H]+ of the ions (1140.50621 + 1329.54306 - 4 = 2466.04927). These results indicate that YW3-56 exists as a pentamer (2466.24467) and the two fragment ions (1140.50621 and 1329.54306) are from the fragmentation of a pentamer.
Figure 5.
Quadrupole-Maldi trandem mass spectrum and fragmentation pattern of YW3-56. a) Locally enlarged peak of the pentamer (2467.24467) and the peaks of the two fragments (1140.50621 and 1329.54306); b) Fragmentation pattern of a pentamer. The formed trimer fragment lost two phenyl groups and the formed dimer fragment got two phenyl groups.
To explain the relationship between the pentamer (2466.24467) and the two fragment ions (1140.50621 and 1329.54306), a fragmentation pattern was postulated and is shown in Figure 5b. This fragmentation pattern suggests that by using a trandem mass condition a pentamer can break into a trimer fragment and a dimer fragment, and the two phenyl groups of the trimer fragment will spontaneously migrate onto the dimer fragment.
The NMR data together with the MS data support that YW3-56 exists as pentamer, and has a tendency to aggregate via a pentamer, i.e. five molecules of YW3-56 are likely involved in the intermolecular assembly.
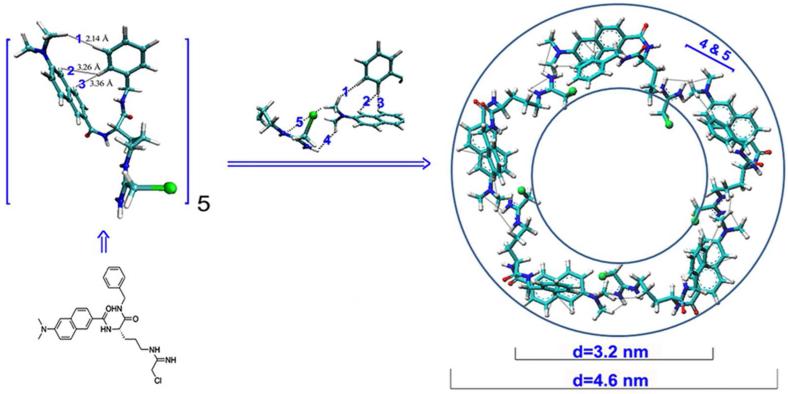
3.5. Computer-assisted assembly mode supporting YW3-56 existing as cyclic pentamers
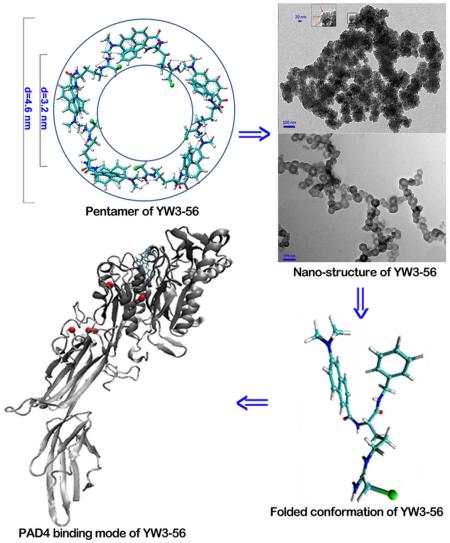
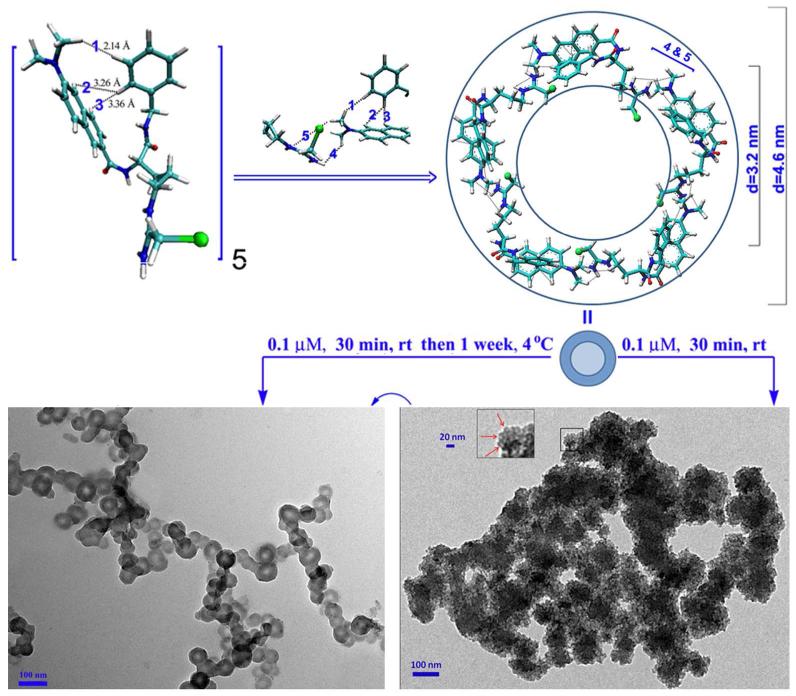
To define the shape of the pentamer the computer-assisted assembly mode was developed. In brief, according to the PAD4 binding conformation of YW3-56, the terminal N of the modified guanido and the naphthyl were arranged in a coplanar manner, and this plane and the plane of the N-benzylformamide have a dihedral angle of 84°. The angle of the modified arginine chain and the 6-dimethylaminonaph-2-yl-carbonyl-amino moiety is 108°, which is the inner angle of a pentagon. Therefore the pentamer was postulated to have a cyclic structure (Figure 6). The length of each side of the pentagon is approximately the total length of the modified arginine chain and the 6-dimethylaminonaph-2-yl-carbonyl-amino moiety, which is 1.9 nm. Therefore, the inner and outer diameters of the circle are approximately 3.2 and 4.6 nm, respectively.
Figure 6.
Computer-assisted assembly mode defined a cyclic pentamer of YW3-56. The 1-3 intramolecular and 4-5 intermolecular interactions observed in the ROESY 2D NMR spectrum were marked.
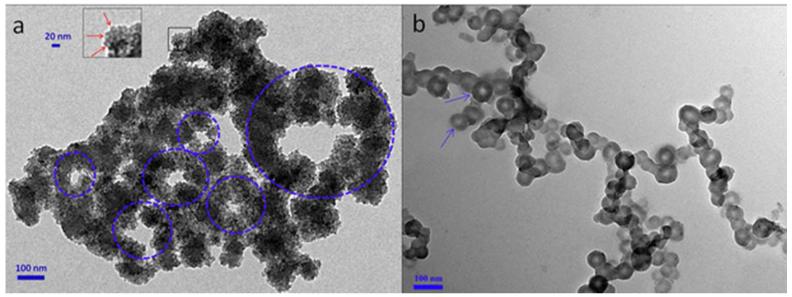
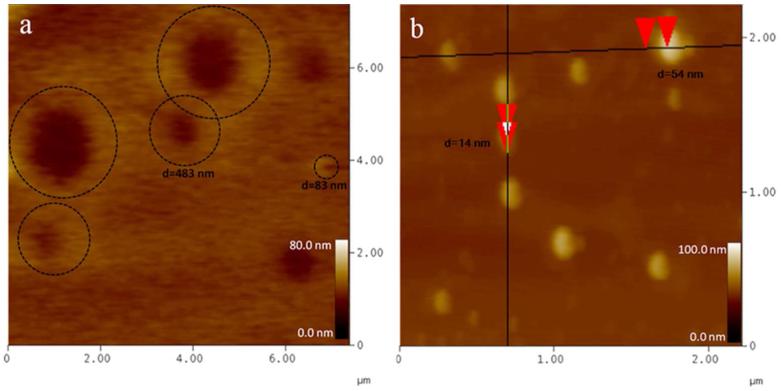
3.6. TEM, SEM and AFM images
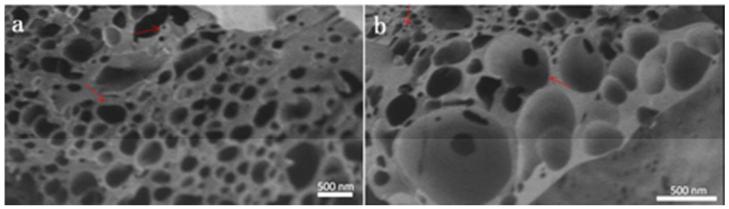
To investigate the nano-structure the TEM, SEM and AFM images of YW3-56 were recorded and are shown in Figure 7-9. The TEM images explore that in ethanol and at room temperature for 30 min the cyclic pentamers of ~5 nm in diameter assembles to nan-rings of 150-600 nm in diameter (Fig. 7a). In ethanol and at 4 °C for one week the cyclic pentamers form nano-capsule of ~50 nm in diameter (Fig. 7b). These observations suggest that the rapidly formed nano-rings can gradually convert to nano-capsules. The SEM images explore that the precipitates resulted from the ethanol solution at room temperature are the aggregator of some nano-rings of 50-500 nm in diameter (Fig. 8a), whereas the precipitates resulted from the ethanol solution at 4 °C are the aggregator of nano-capsules of 88-529 nm in diameter (Fig. 8b). The AFM images explore that the precipitates resulted from the ethanol solution at room temperature are the aggregator of nano-rings of 83-762 nm in diameter (Fig. 9a), whereas the precipitates resulted from the ethanol solution at 4 °C are the aggregator of nano-capsules of 14-54 nm in diameter (Fig. 9b).
Figure 7.
TEM images of YW3-56, in ethanol at room temperature kept for 30 min (a), and in ethanol at 4 °C kept for one week (b). The red arrows in the magnified area refer to the cyclic pentamers (a).
Figure 9.
AFM images of YW3-56, in ethanol at room temperature kept for 30 min (a) and in ethanol at 4 °C kept for one week (b). The ring-like aggregators in panel a are circled by dashed lines. The red triangles in panel b measure the vertical distances of the nano-particles.
Figure 8.
SEM images of YW3-56, in ethanol at room temperature kept for 30 min (a) and in ethanol at 4 °C kept for one week (b). The red arrows refer to the smaller and larger nano particles.
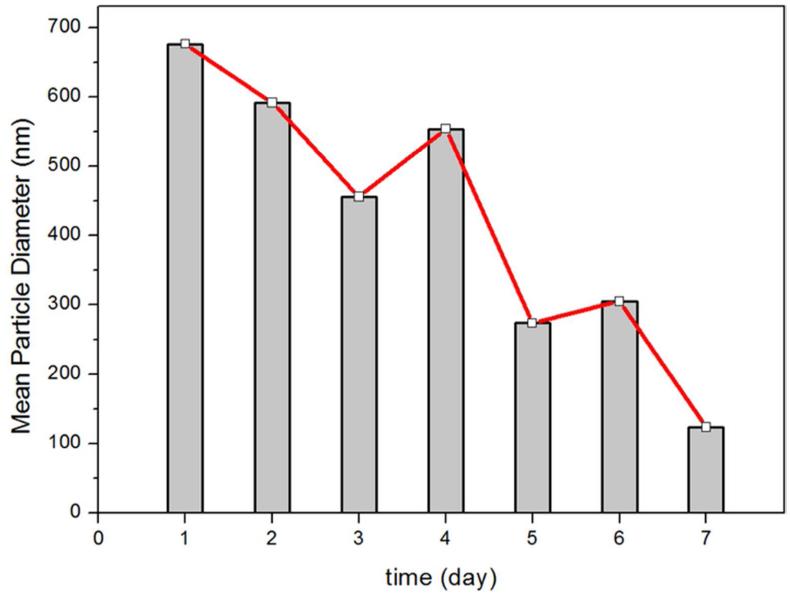
To examine the stability of the nano-capsules the mean particle diameter measurements were performed with a Malvern’s Zeta Sizer (Nano-ZS90) with DTS (Nano) Program by using the standard procedure, and the result is shown in Figure 10. The data demonstrate that the mean particle diameter tends to decrease when the ethanol solution of YW3-56 is kept at 4 °C for longer. It changed from ~680 nm in day 1 to ~120 nm in day 7. This means that the nano-capsules of YW3-56 aggregates tighter as time goes by.
Figure 10.
The mean particle diameter of YW3-56 in ethanol at 4 °C as time goes by.
3.7. The interactions in the formation of nano-structures
The ROESY 2D NMR spectrum (Figure 2) also provides the evidences for the physical and chemical mechanisms of the cyclic pentamers forming the nano-rings and the nano-capsules. The cross-peak 6 is for the H of the NCH3 of one cyclic pentamer and the H of CH2Cl of another cyclic pentamer, the cross-peak 7 is for the 4-H of the naphthyl of one cyclic pentamer and the H of the NCH3 of another cyclic pentamer, the cross-peak 8 is for the H of the CH2Cl of one cyclic pentamer and the 1-H of the naphthyl of another cyclic pentamer, and the cross-peak 9 is for the H of the NCH3 of one cyclic pentamer and the 1-H of the naphthyl of another cyclic pentamer. Thus there are 4 kinds of interactions between the cyclic pentamers. These interactions drive YW3-56 forming nano-rings and nano-capsules.
3.8. Structural determinants of higher anti-cancer efficacy of YW3-56 than benzoyl-L-Arg
It was documented that the in vivo anti-cancer efficacy of YW3-56 was significantly higher than that of benzoyl-L-Arg.26 In the molecular docking of YW3-56 towards the active site pocket of PAD4, though the modified arginine chain of YW3-56 and benzoyl-L-Arg have similar binding mode, the 6-dimethylaminonaph-2-yl-carbonyl-amino moiety of YW3-56 has additional interactions such as the hydrogen bonds with residues Arg372, Arg374, Arg441 and Asp350. Thus a rational understanding should be that these additional interactions benefit the PAD4 inhibition efficacy of YW3-56, which in turn benefits the anti-cancer efficacy of YW3-56.
CONCLUSIONS
PAD1-4 and 6 convert arginine residues to citrullines and have tissue and substrate specificity.27,28 Of them, PAD4 possesses a nuclear localization signal and citrullinates many substrates including histones, p300/CBP, NPM, ING4 and nuclear lamin C to exert various functions.29-33 The pathology studies link PAD4 to the etiology of cancers of human being.34-44 For discovering small molecule inhibitors of PAD4 the knowledge of the folded conformation, cyclic pentamer, nano-structure and PAD4 binding mode of YW3-56 is of clinical importance. The π-π stacking is the driven force of YW3-56 forming two folded conformations 1 and 2. The active site pocket of PAD4 prefers the folded conformation 2. The intermolecular interactions direct five molecules of YW3-56 to form cyclic pentamer. The intermolecular interactions between the cyclic pentamers drive them to rapidly forming nano-rings. The nano-rings then gradually assemble to the nano-capsules. These knowledge are summarized in Figure 11.
Figure 11.
The relationships between the folded conformation 2, the cyclic pentamer, the nano rings and the nano-capsules.
Numerous examples of macromolecular self-assembly, such as self-assembly of layer-by-layer,45-47 self-assembly of different guest substrates on the surfaces of bulk natural cellulose substances,48 self-assembly of block copolymers,49 self-assembly of dendronized non-planar conjugated molecules,50 self-assembly of hemoproteins,51 were previously reported. The formation of a macromolecular self-assembly system usually needs multiple components, special fabrication procedure, and solid surface or scaffold. In contrast to a macromolecular self-assembly, our studies show that a small molecular self assembly system can be fabricated in significantly simple condition.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by Engineering Research Center of Endogenous Prophylactic of Ministry of Education of China, by PHR (IHLB), by the National Natural Science Foundation (81172930, 81273379 and 81202412) and by Special Project (2011ZX09302-007-01) of China, as well as by a NCI/NIH grant R01 CA136856 to Y.M.W.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Description of the synthetic route, NMR spectra, conformations in different solvent and additional TEM image of YW3-56 were included. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Lee O-S, Prytkova TR, Schatz GC. Using DNA to Link Gold Nanoparticles, Polymers and Molecules: A Theoretical Perspective. J. Phys. Chem. Lett. 2010;1:1781–1788. doi: 10.1021/jz100435a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Kemling JW, Qavi AJ, Bailey RC, Suslick KS. Nanostructured Substrates for Optical Sensing. J. Phys. Chem. Lett. 2011;2:2934–2944. doi: 10.1021/jz201147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gagner JE, Shrivastava S, Qian X, Dordick JS, Siegel RW. Engineering Nanomaterials for Biomedical Applications Requires Understanding the Nano-bio Interface: A Perspective. J. Phys. Chem. Lett. 2012;3:3149–3158. doi: 10.1021/jz301253s. [DOI] [PubMed] [Google Scholar]
- (4).Berweger S, Atkin JM, Olmon RL, Raschke MB. Adiabatic Tip-plasmon Focusing for Nano-raman Spectroscopy. J. Phys. Chem. Lett. 2010;1:3427–3432. [Google Scholar]
- (5).Meng F, Zhong Z. Polymersomes Spanning from Nano- to Microscales: Advanced Vehicles for Controlled Drug Delivery and Robust Vesicles for Virus and Cell Mimicking. J. Phys. Chem. Lett. 2011;2:1533–1539. [Google Scholar]
- (6).Kumari G, Narayana C. New Nano Architecture for SERS Applications. J. Phys. Chem. Lett. 2012;3:1130–1135. doi: 10.1021/jz3001344. [DOI] [PubMed] [Google Scholar]
- (7).Luan B, Zhou R. Nanopore-based Sensors for Detecting Toxicity of a Carbon Nanotube to Proteins. J. Phys. Chem. Lett. 2012;3:2337–2341. doi: 10.1021/jz3007832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Odom TW, You E-A, Sweeney CM. Multiscale Plasmonic Nanoparticles and the Inverse Problem. J. Phys. Chem. Lett. 2012;3:2611–2616. doi: 10.1021/jz300886z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Anthony SP, Draper SM. Nano/Microstructure Fabrication of Functional Organic Material: Polymorphic Structure and Tunable Luminescence. J. Phys. Chem. C. 2010;114:11708–11716. [Google Scholar]
- (10).Sood R, Iojoiu C, Espuche E, Gouanvé F, Gebel G, Mendil-Jakani H, Lyonnard S, Jestin J. Proton Conducting Ionic Liquid Doped Nafion Membranes: Nano-Structuration, Transport Properties and Water Sorption. J. Phys. Chem. C. 2012;116:24413–24423. [Google Scholar]
- (11).Marcelo G, Muñoz-Bonilla A, Fernández-García M. Magnetite–Polypeptide Hybrid Materials Decorated with Gold Nanoparticles: Study of Their Catalytic Activity in 4-Nitrophenol Reduction. J. Phys. Chem. C. 2012;116:24717–24725. [Google Scholar]
- (12).Mir M, Tahirbegi IB, Valle-Delgado JJ, Fernàndez-Busquets X, Samitier J. In vitro Study of Magnetite-amyloid β Complex Formation. Nanomed-nanotechnol. 2012;8:974–980. doi: 10.1016/j.nano.2011.11.010. [DOI] [PubMed] [Google Scholar]
- (13).Mishra B, Patel BB, Tiwari S. Colloidal Nanocarriers: A Review on Formulation Technology, Types and Applications toward Targeted Drug Delivery. Nanomed-nanotechnol. 2010;6:9–24. doi: 10.1016/j.nano.2009.04.008. [DOI] [PubMed] [Google Scholar]
- (14).Khadka DB, Haynie DT. Protein- and Peptide-based Electrospun Nanofibers in Medical Biomaterials. Nanomed-nanotechnol. 2012;8:1242–1262. doi: 10.1016/j.nano.2012.02.013. [DOI] [PubMed] [Google Scholar]
- (15).Rangnekar A, Zhang AM, Li SS, Bompiani KM, Hansen MN, Gothelf KV, Sullenger BA, LaBean TH. Increased Anticoagulant Activity of Thrombin-binding DNA Aptamers by Nanoscale Organization on DNA Nanostructures. Nanomed-nanotechnol. 2012;8:673–681. doi: 10.1016/j.nano.2011.08.011. [DOI] [PubMed] [Google Scholar]
- (16).Kawasaki ES, Player TA. Nanotechnology, Nanomedicine, and the Development of New, Effective Therapies for Cancer. Nanomed-nanotechnol. 2005;1:101–109. doi: 10.1016/j.nano.2005.03.002. [DOI] [PubMed] [Google Scholar]
- (17).Sundaram P, Wower J, Byrne ME. A Nanoscale Drug Delivery Carrier Using Nucleic Acid Aptamers for Extended Release of Therapeutic. Nanomed-nanotechnol. 2012;8:1143–1151. doi: 10.1016/j.nano.2012.01.010. [DOI] [PubMed] [Google Scholar]
- (18).Li L, Cui G, Zhao M, Wang Y, Wang H, Li W, Peng S. Assembly of β-Cyclo-dextrin with 3S-Tetrahydro-β-carboline-3-carboxylic Acid and Self-assembly of 6-(3S-Carboline-3-carboxylaminoethylamino)-6-deoxy-β-cyclodextrin: Approaches to Enhance Anti-oxidation Stability and Anti-thrombotic Potency. J. Phy. Chem. B. 2008;112:12139–12147. doi: 10.1021/jp8025495. [DOI] [PubMed] [Google Scholar]
- (19).Ren X, Cui G, Zhao M, Wang C, Peng S. Coordination of Thrombolytic Pro- Ala-Lys Peptides with Cu (II): Leading to Nano-scale Self-assembly, Increase of Thrombolytic Activity and Additional Vasodilation. J. Phys. Chem. B. 2008;112:8174–8180. doi: 10.1021/jp800645g. [DOI] [PubMed] [Google Scholar]
- (20).Abelein A, Lang L, Lendel C, Gräslund A, Danielsson J. Transient small molecule interactions kinetically modulate amyloid β peptide self-assembly. FEBS Lett. 2012;586:3991–3995. doi: 10.1016/j.febslet.2012.09.035. [DOI] [PubMed] [Google Scholar]
- (21).Sood A, Abid M, Hailemichael S, Foster M, Török B, Török M. Effect of Chirality of Small Molecule Organofluorine Inhibitors of Amyloid Self-Assembly on Inhibitor Potency. Bioorg. Med. Chem. Lett. 2009;19:6931–6934. doi: 10.1016/j.bmcl.2009.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kühnle A. Self-assembly of organic molecules at metal surfaces. Curr. Opin. Colloid Int. 2009;14:157–168. [Google Scholar]
- (23).Matson JB, Zha RH, Stupp SI. Peptide Self-Assembly for Crafting Functional Biological Materials. Curr. Opin. Solid St. M. 2011;15:225–235. doi: 10.1016/j.cossms.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wang Y, Wu J, Kang G, Zhao M, Gui L, Li N, Peng L, Zhang X, Li L, Peng S. Novel nano-materials, RGD-tetrapeptide -modified 17β-amino-11α-hydroxyandrost-1,4-diene-3-one: synthesis, self-assembly based nano images and in vivo anti-osteoporosis evaluation. J. Mater. Chem. 2012;22:4652–4659. [Google Scholar]
- (25).Wang Y, Li P, Wang S, Hu J, Chen XA, Wu J, Fisher M, Oshaben K, Zhao N, Gu Y, et al. Anticancer PAD Inhibitors Regulate the Autophagy Flux and the Mammalian Target of Rapamycin Complex 1 Activity. J. Biol. Chem. 2012;17:25941–25953. doi: 10.1074/jbc.M112.375725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a Growing Family of Citrullinating Enzymes: Genes, Features and Involvement in Disease. Bioessays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- (27).Darrah E, Rosen A, Giles JT, Andrade F. Peptidylargininedeiminase 2, 3 and 4 Have Distinct Specificities Against Cellular Substrates: Novel Insights into Autoantigen Selection in Rheumatoid Arthritis. Ann. Rheum. Dis. 2012;71:92–98. doi: 10.1136/ard.2011.151712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Nakashima K, Hagiwara T, Ishigami A, Nagata S, Asaga H, Kuramoto M, Senshu T, Yamada M. Molecular Characterization of Peptidylargininedeiminase in HL-60 Cells Induced by Retinoic Acid and 1-Alpha, 25-Dihydroxyvitamin D (3) J. Biol. Chem. 1999;274:27786–27792. doi: 10.1074/jbc.274.39.27786. [DOI] [PubMed] [Google Scholar]
- (29).Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, et al. Human PAD4 Regulates Histone Arginine Methylation Levels Via Demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- (30).Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of Coactivator Complex Assembly and Function by Protein Arginine Methylation and Demethylimination. Proc. Natl. Acad. Sci. USA. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Guo Q, Fast W. Citrullination of Inhibitor of Growth 4 (ING4) by Peptidylargininedeminase 4 (PAD4) Disrupts the Interaction Between ING4 and p53. J. Biol. Chem. 2011;286:17069–17078. doi: 10.1074/jbc.M111.230961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Tanikawa C, Espinosa M, Suzuki A, Masuda K, Yamamoto K, Tsuchiya E, Ueda K, Daigo Y, Nakamura Y, Matsuda K. Regulation of Histone Modification and Chromatin Structure by the p53-PADI4 Pathway. Nat. Commun. 2012;3:676. doi: 10.1038/ncomms1676. [DOI] [PubMed] [Google Scholar]
- (33).Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono M, et al. Functional Haplotypes of PADI4, Encoding Citrullinating Enzyme Peptidylargininedeiminase 4, Are Associated With Rheumatoid Arthritis. Nat. Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- (34).Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. Increased PADI4 Expression in Blood and Tissues of Patients With Malignant Tumors. BMC Cancer. 2009;9:40. doi: 10.1186/1471-2407-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wang L, Chang X, Yuan G, Zhao Y, Wang P. Expression of Peptidylargininedeiminase Type 4 in Ovarian Tumors. Int. J. Biol. Sci. 2010;6:454–464. doi: 10.7150/ijbs.6.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Chang X, Hou X, Pan J, Fang K, Wang L, Han J. Investigating the Pathogenic Role of PADI4 in Oesophageal Cancer. Int. J. Biol. Sci. 2011;7:769–781. doi: 10.7150/ijbs.7.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR, Gilmour DS, Wang Y. Regulation of p53 Target Gene Expression by Peptidylargininedeiminase 4. Mol. Cell Biol. 2008;28:4745–4758. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Li P, Wang D, Yao H, Doret P, Hao G, Shen Q, Qiu H, Zhang X, Wang Y, Chen G. Coordination of PAD4 and HDAC2 in the Regulation of p53-target Gene Expression. Oncogene. 2010;29:3153–3162. doi: 10.1038/onc.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Kruse JP, Gu W. Modes of p53 Regulation. Modes of p53 Regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced Modulator of Autophagy, Is Critical for Apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- (41).Tang Y, Luo J, Zhang W, Gu W. Tip60-Dependent Acetylation of p53 Modulates the Decision between Cell-cycle Arrest and Apoptosis. Mol. Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- (42).Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two Mammalian MOF Complexes Regulate Transcription Activation by Distinct Mechanisms. Mol. Cell. 2009;36:290–301. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Vousden KH, Prives C. Blinded by The Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- (44).Tanikawa C, Ueda K, Nakagawa H, Yoshida N, Nakamura Y, Matsuda K. Regulation of Protein Citrullination Through p53/PADI4 Network in DNA Damage Response. Cancer Res. 2009;69:8761–8769. doi: 10.1158/0008-5472.CAN-09-2280. [DOI] [PubMed] [Google Scholar]
- (45).Sun Q, Tong Z, Wang C, Ren B, Liu X, Zeng F. Charge density threshold for LbL self-assembly and small molecule diffusion in polyelectrolyte multilayer films. Polymer. 2005;46:4958–4966. [Google Scholar]
- (46).Huang J, Gu Y. Self-assembly of various guest substrates in natural cellulose substances to functional nanostructured materials. Curr. Opin. Colloid Int. 2011;16:470–481. [Google Scholar]
- (47).de Villiers MM, Otto DP, Strydom SJ, Lvov YM. Introduction to nanocoatings produced by layer-by-layer (LbL) self-assembly. Adv. Drug Deliver. Rev. 2011;63:701–715. doi: 10.1016/j.addr.2011.05.011. [DOI] [PubMed] [Google Scholar]
- (48).Deshmukh PK, Ramani KP, Singh SS, Tekade AR, Chatap VK, Patil GB, Bari SB. Stimuli-sensitive layer-by-layer (LbL) self-assembly systems: Targeting and biosensory applications. J. Control. Release. 2013;166:294–306. doi: 10.1016/j.jconrel.2012.12.033. [DOI] [PubMed] [Google Scholar]
- (49).Ho R-M, Chiang Y-W, Lin S-C, Chen C-K. Helical Architectures from Self-assembly of Chiral Polymers and Block Copolymers. Prog. Polym. Sci. 2011;36:376–453. [Google Scholar]
- (50).Yang Y, Miao X, Liu G, Xu L, Wu T, Deng W. Self-assembly of dendronized non-planar conjugated molecules on a HOPG surface. Appl. Surf. Sci. 2012;263:73–78. [Google Scholar]
- (51).Oohora K, Onoda A, Hayashi T. Supramolecular assembling systems formed by heme-heme pocket interactions in hemoproteins. Chem. Commun. 2012;48:11714–11726. doi: 10.1039/c2cc36376c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.