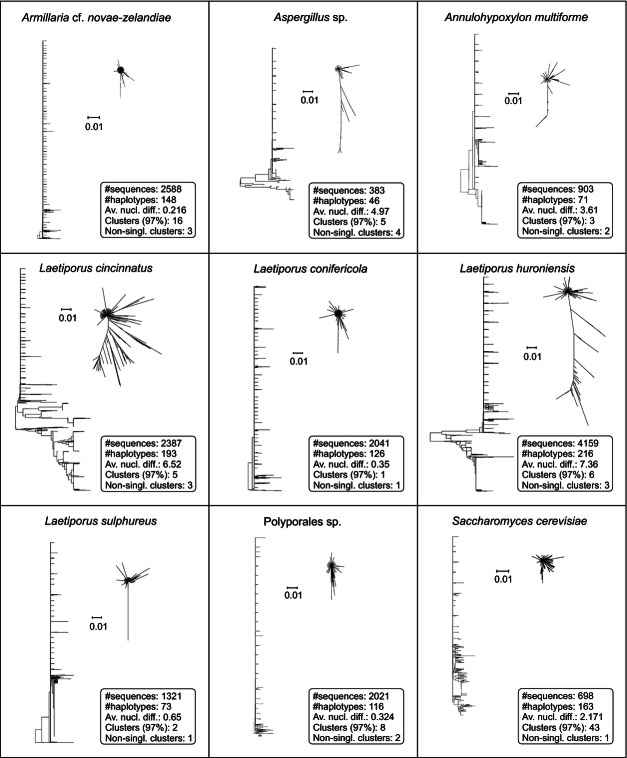
Figure 2.
Neighbor-joining trees illustrating sequence variation in the ITS1 alignments in species with varying levels of molecular variation. Armillaria cf. novae-zelandiae, Aspergillus sp., Annulohypoxylon multiforme, Laetiporus cincinnatus, L. huroniensis, Polyporales sp., and Saccharomyces cerevisiae are included as species with high levels of variation, whereas Laetiporus conifericola and L. sulphurensis are included as typical examples representing species with lower levels of variation. Both midpoint rooted and unrooted trees are shown for all taxa. Similar scales are used across all trees to enable direct comparisons. We hypothesize that the star-shaped unrooted trees observed for Armillaria cf. novae-zelandiae, L. conifericola, L. sulphurensis, Polyporales sp., and Saccharomyces cerevisiae are due to PCR and sequencing errors, whereas the more complex trees for Aspergillus sp., Annulohypoxylon multiforme, L. cincinnatus, and L. huroniensis are due to intragenomic variation.