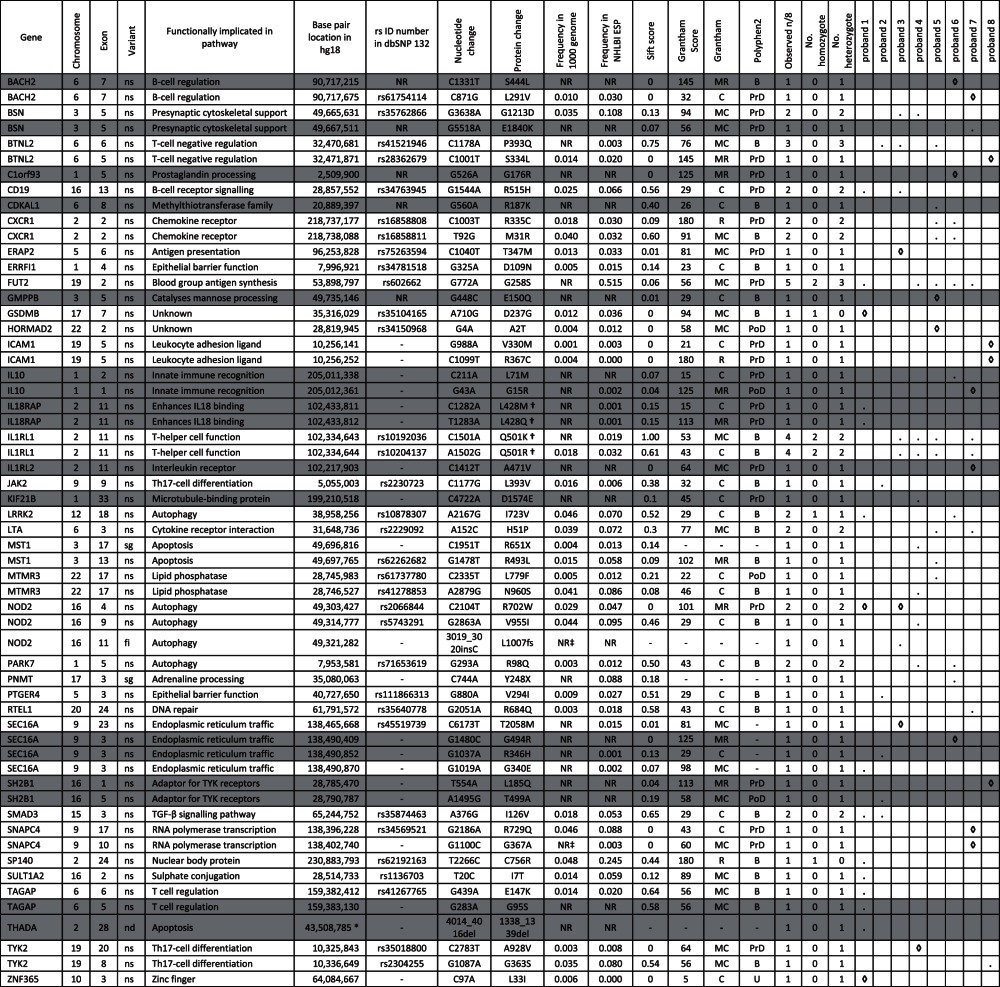
Table 2.
Characterisation of non-synonymous, stopgain and indel variants with an alternative allele frequency of <0.05 or not reported in 1000 genomes across 39 known IBD genes
|
Novel variants are shown in grey.
Where a specific variant is present in a proband, this is indicated by a dot (.).
Where a specific variant is present in a proband and has a SIFT score of <0.05 this is indicated by a ◊.
*Indicates the first bp location of a 3-bp deletion.
†Indicates a dinucleotide variant (that for IL18RAP results in a codon change from CTG>AAG, resulting in p.L428K amino acid change).
‡NR indicates variants that despite not being reported in dbSNP132 or 1000 genomes, are reported in dbSNP129 or seen in our in-house reference exomes and are therefore not characterised as novel.
B, benign; C, conservative; fi, frameshift insertion; MC, moderately conservative; MR, moderately radical; nd, non-frameshift deletion; NR, not reported; ns, non-synonymous; PoD, possibly damaging; PrD, probably damaging; R, radical; sg, stopgain; U, unknown.