Figure 1.
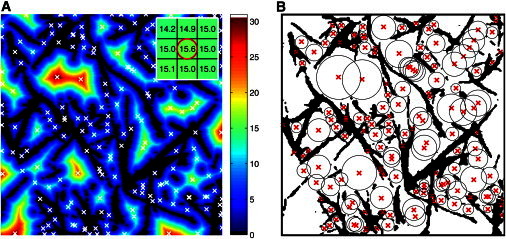
Simplified bubble analysis of the pore space of a random biopolymer network in two steps. (A) First, the Euclidean distance map (EDM) of the fluid space of the network structure is computed (shades of gray (colors online) indicate the distance of each fluid pixel to the nearest fiber pixel). (Black) Fibers. Second, the local maxima of the EDM (white crosses) determine the centers of all 2D bubbles. To avoid bubbles of similar size in close proximity, the EDM was smoothed with a 5 × 5 Gaussian kernel with a sigma of one before the local maxima were determined. (Inset) A local maximum of the EDM (red circle) is a pixel whose eight neighbors all have smaller values. (B) Resulting 2D bubbles (black circles) fit into the pore zones of the fiber structure (black). (Red crosses) Bubble centers.
