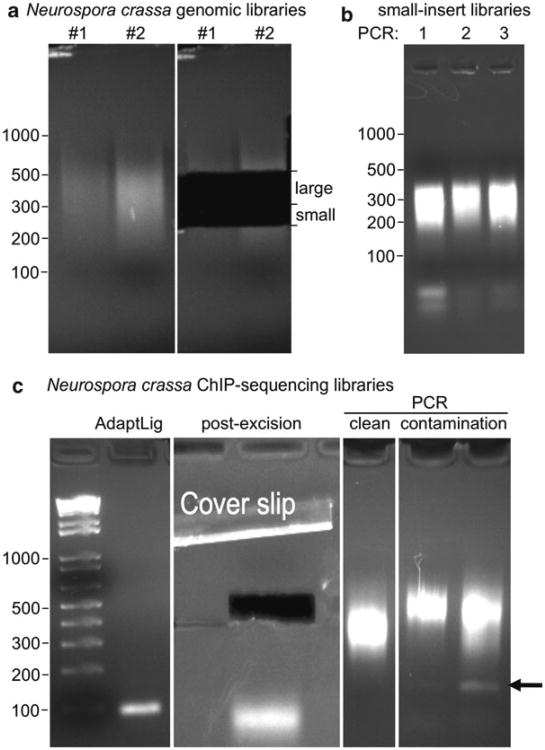
Fig. 3.
Preparation of genomic and ChIP paired-end sequencing libraries. (a) Neurospora crassa genomic DNA was sonicated with a tip sonicator (see Note 5), and libraries were generated as described. Two replicates were done in parallel and loaded on a 2% NuSieve agarose gel for DNA isolation (left panel). Two fractions were removed from the gel (250–325 bp, “small”; 325–500 bp, “large”). We do this to insure that we have library material for future PCRs even in cases when library amplifications fail or other unforeseen difficulties arise. (b) Three parallel PCR amplifications of the small insert libraries shown in (a) (5 μL of 35 μL were loaded). Note the presence of adapter and PCR primer bands. (c) Example of construction of ChIP-seq libraries. After ligation of adapters (“AdaptLig”) the library is invisible on the gel (but note the presence of adapter bands below 100 bp). After excision of the bands and extraction of DNA from the gel (Qiagen gel extraction kit), PCR reactions that are puri fi ed (Qiagen PCR puri fi cation kit) typically yield slightly smeary library bands (“clean”) but no bands <200 bp. It is important to adjust the adapter:DNA ratio to ∼10:1. In cases of great adapter excess, which can be a problem with ChIP-seq library generation, spurious bands are observed. Paired-end adapters run at ∼160 bp and single-end adapters run at ∼110 bp (“contamination,” see arrow). We would not subject the library in the right-most lane to sequencing as the amount of contamination present will significantly reduce the number of useable reads, because most reads will be adapter sequence instead of insert.