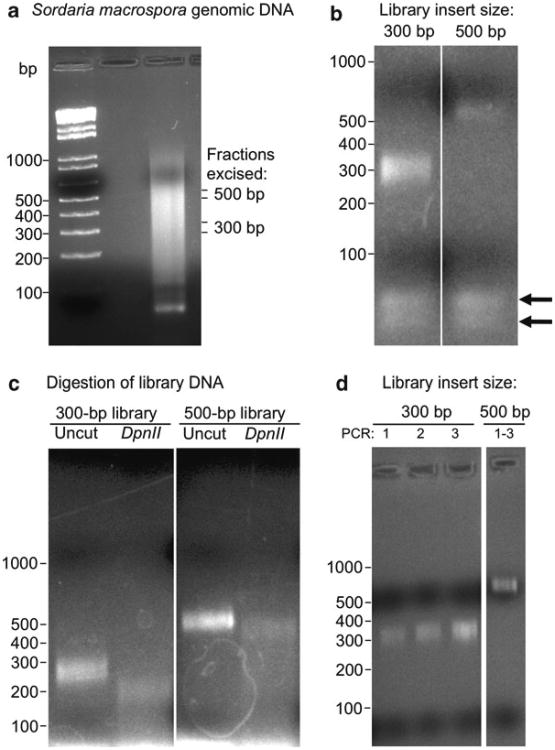
Fig. 4.
Preparation of paired-end sequencing libraries from Sordaria macrospora. (a) Genomic DNA (10 μg in 450 μL TE) was prepared as described in ref. (11) and sheared by sonication with a Branson 450 Sonifier with microtip (duty cycle 80%, output 1.2; five cycles of 10 s pulses, interrupted by 30 s rest on ice). Sheared DNA was concentrated (Qiagen PCR purification kit) and separated on a 1.2% agarose gel. Gel slices of ∼300 and ∼500 bp fragments were isolated and purified (Qiagen Gel extraction kit, see Subheading 3.4, step 5). (b) Paired-end libraries were constructed as described in the methods from the 300- and 500-bp fractions and amplified with 12 cycles of PCR, yielding barely visible bands. Note presence of adapters and PCR primers below the 100 bp marker. (c) Test for library complexity. One simple test to show that libraries contain predominantly genomic DNA and are not enriched for adapter dimers or other spurious overamplified bands is to digest the DyNAzyme-amplified libraries with DpnII. This cleaves most of the adapter off the inserts. If genomic DNA has been cloned it will result in a smear (similar to genomic DNA digested for Southern blots). If only low complexity samples have been cloned in the library, sharp banding is observed instead of the smear and the library is unsuitable for further processing. (d) Libraries were amplified with 18 cycles of PCR with Phusion Flash High-Fidelity polymerase and purified (Qiagen PCR purification kit; see three 300-bp library samples). We typically pool three independent 25 μmL PCR reactions before purification and run 5 μL of 35 μL to show that only expected bands are obtained. Note the absence of the adapters or PCR primers and primer dimers (compare to (b)).