Abstract
Conformal radiotherapy planning needs accurate delineations of the critical structures. Atlas-based segmentation has been shown to be very efficient to delineate brain structures. It would therefore be very interesting to develop an atlas for the head and neck region where 7 % of the cancers arise. However, the construction of an atlas in this region is very difficult due to the high variability of the anatomies. This can generate segmentation errors and over-segmented structures in the atlas. To overcome this drawback, we present an alternative method to build a template locally adapted to the patient’s anatomy. This is done first by selecting in a database the images that are the most similar to the patient on predefined regions of interest, using on a distance between transformations. The first major contribution is that we do not compute every patient-to-image registration to find the most similar image, but only the registration of the patient towards an average image. This method is therefore computationally very efficient. The second major contribution is a novel method to use the selected images and the predefined regions to build a Frankenstein’s creature” for segmentation. We present a qualitative and quantitative comparison between the proposed method and a classical atlas-based segmentation method. This evaluation is performed on a subset of 58 patients among a database of 105 head and neck CT images and shows a great improvement of the specificity of the results.
1 Introduction
Conformal radiotherapy allows to precisely target the tumor while keeping an acceptable level of irradiation on neighboring critical structures. This however requires to accurately locate the tumor and the organs at risk in order to determine the best characteristics of the irradiation beams. This delineation task is usually done manually and is therefore very long and not reproducible.
To solve this problem, atlas-based segmentation has been shown to produce accurate and automatic segmentations of the brain [1], allowing to take into account easily the relative positions of the structures. The construction of an atlas for other regions such as the head and neck region, where 7 % of the cancers arise, is therefore of great interest [2, 3]. Methods have been presented for the construction of an unbiased average model from an image dataset such as [4, 5]. We presented in [6] a method to build a symmetric atlas from a database of images manually delineated following the guidelines in [7]. However, anatomical variability is very high in the head and neck region. An average atlas may therefore be very different from the patient, leading to registration discrepancies. Moreover, this variability may cause the mean contours to be too large in the atlas yielding over-segmentations.
To overcome these drawbacks, methods have been presented towards the creation of atlases whose anatomy is adapted to the patient [8, 9]. First, Blezek et al. [8] presented an interesting approach to cluster a database into several atlases representing homogeneous sub-populations. However, the selection of the most adequate atlas with respect to a given patient is not addressed. Another method [9] has been introduced to select the most similar images to the patient by comparing a similarity measure between each database image and the patient. This method is however computationally expensive, requiring to register all the database images on the patient. Moreover, a local comparison of the images is more adapted in our case, as our database consists of manually delineated patients who present pathologies that may corrupt a global comparison.
In this paper, we present the development of an atlas locally adapted to the patient to get more precise delineations than with an average atlas. To this end, we first present a new and efficient method to select the most similar images to the patient on predefined regions. Each most similar sample is defined as the one that needs the smallest local deformations to be registered on the patient. These images are then combined into a template image, as an analogy to Frankenstein’s creature [10], and used for segmentation.
We first present our approach to select the image that is the most similar to the patient P to delineate on a given region. We then focus on the combination of the local templates selected into one single template for delineation. Finally, we show qualitative and quantitative results on a database of 105 head and neck CT images, showing a great improvement of the specificity of the results.
2 Method
In this section, we present an efficient method to compute a template that is similar to the patient P on predefined regions Rl, based on the following steps:
-
–
Construction of an average atlas M from the database images (pre-computed)
-
–
Non linear registration of the patient P to delineate on M
-
–
Selection of the most similar image Ĩl for each local region Rl
-
–
Computation of the anatomy M̃ and segmentations from the set of Ĩl
2.1 Selection of the Locally Most Similar Images to a Patient
For each region of interest Rl, we select the most similar image among our database to the patient to delineate. It is defined as the one that is the “less” deformed to be non linearly registered on the patient. We denote by TB←A the transformation linking two images A and B, so that B can be resampled on A (i.e. A ≈ B○TB ←A). The selection for a given region Rl is then based on a comparison of the non linear transformations TIj←P to bring each image Ij of the database on P, i.e. the most similar image is defined as: Ĩl = arg minIj dRl (Ij, P) = arg minIj dRl (TIj←P, Id), where dRl will be defined later on. However, this type of comparison is computationally very expensive as it requires to perform all the registrations between P and the images Ij for each patient to segment. To perform an efficient selection, we therefore use an intermediate image: an average atlas M pre-computed from the database using [6].
From the average atlas construction, we obtain for each image Ij an affine transformation AIj←M and a non linear transformation TIj←M bringing it on the average image M. Moreover, when registering P on M, another non linear transformation TP←M is computed. The key hypothesis is then to assume that TIj←P can be approximated by TIj←M ○ . This hypothesis presents many advantages. First, the regions of interest Rl can be defined once and for all on the atlas image M. Moreover, this can be done very easily thanks to the average segmentations available on the atlas. Also, the similarity between P and Ij, dRl (TIj←P, Id), can be approximated using the following equation:
| (1) |
where i corresponds to the voxels of the dense transformation and dRl is the Log-Euclidean distance on diffeomorphisms [11] between the identity transformation and TIj←M ○ . Using our hypothesis, we need to perform only one non linear registration between M and P to select the locally most similar images Ĩl for all regions Rl, therefore reducing drastically the computation time.
2.2 Piecewise Most Similar Atlas Construction
We now focus on the computation of a template for segmentation from the selected images Ĩl and the regions Rl. This template, similar to the Frankenstein’s creature [10], is built by iterating over the following steps to combine the images:
-
–
Registration of the images Ĩl on the average image at iteration k : M̃k
-
–
Compute the new average image Mk+1, based on the regions Rl,k
-
–
Compute an average transformation T̄k from the transformations TĨl←M̃k
-
–
Apply to Mk+1 to get the new reference M̃k+1 = Mk+1 ○
-
–
Update the regions of interest by applying to Rl,k : Rl,k+1 = Rl,k ○
This algorithm can be seen as an extension of [4] to the construction of an atlas where images have spatially varying weights, depending on the regions Rl. In contrast, Guimond et al. consider implicitly that all images have equal and spatially constant weights (1/N for each image if N images are averaged). The final step is then to associate a set of segmentations to this anatomy. This is done by transforming the manual segmentations of the image Ĩl present in the region Rl onto M̃, using the transformations obtained in the construction process, and ensuring that no overlap exists between the final structures.
Average Image Computation
To compute Mk+1 from the images Ĩl registered on M̃k, we first need to define the spatial extensions of each region at iteration k : Rl,k. This will allow to use, on each Rl,k, only the corresponding Ĩl and to interpolate between the regions. The weight functions w̄l,k(x) are computed in three steps: locally erode the regions Rl,k using the method presented in [12] to ensure a minimal distance between them, compute the inverse of the minimal distance to each Rl,k: wl,k(x) = 1/(1 + αdist(x, Rl,k)), and normalize the .
The images Ĩl are then aligned onto M̃k, first globally resulting in affine transformations AĨl←M̃k, and then non linearly producing dense transformations TĨl←M̃k. These transformations and the w̄l,k(x) are then used to compute Mk+1:
| (2) |
Residual Deformation Computation
Similarly to [4], the next step consists in averaging the TĨl←M̃k into a transformation T̄k and apply its inverse to Mk+1 to get the new reference M̃k+1 = Mk+1 ○ . However, we are averaging transformations using spatially variable weights w̄l,k(x). To take this into account and ensure that T̄k is a diffeomorphism, we introduce a generalization of the Log-Euclidean (LE) polyaffine transformation to diffeomorphisms, as suggested in [13]. The polydiffeomorphism construction is based on the LE framework for diffeomorphisms [11], allowing to compute operations easily while staying on the manifold of diffeomorphisms. T̄k is then built by integrating between time 0 : x(0) = x and time 1 : x(1) = T̄k(x) the following Ordinary Differential Equation (ODE): . Similarly to the LE polyaffine framework, T̄k and are expressed respectively as the exponential of the right hand side of the ODE, and the exponential of its opposite.
3 Evaluation Methodology
To evaluate our method, we have used a database of 105 CT images of patients delineated for head and neck radiotherapy following the guidelines provided in [7]. Segmented structures included 12 structures: lymph nodes II, III and IV, parotids and sub-mandibular glands on each side as well as the spinal cord and the brainstem. On this database, we have repeated a Leave-One-Out approach, each time picking out one patient from the dataset of images. The average atlas is then built from the remaining images. We used this framework to compare two segmentation methods: the average atlas-based segmentation, and the locally most similar image based segmentation. The Frankenstein’s creature was built by defining on the average atlas a region of interest for each structure. This gives a total of 12 selected images, combined together into a single composite patient. This image is then registered on the left-out patient to get its segmentation.
The results were then compared to the manual delineations of the left-out patient using two voxel-based overlap measures: sensitivity (rate of true detection of the structure) and specificity (rate of true detection of the background). As they were delineated for radiotherapy, some structures were not available. We have therefore used the Leave-One-Out evaluation on a subset of 58 patients which had 8 or more manual delineations. Finally, the separation between some structures (lymph nodes, brainstem and spinal cord) are made on an arbitrary axial plane, based on possibly moving anatomical landmarks. This may lead to errors in the separations of the automatic segmentations and artificially low quality measures for all methods. We addressed this by evaluating together the brainstem and spinal cord, and by grouping the lymph nodes on each side.
4 Results
4.1 Qualitative Evaluation
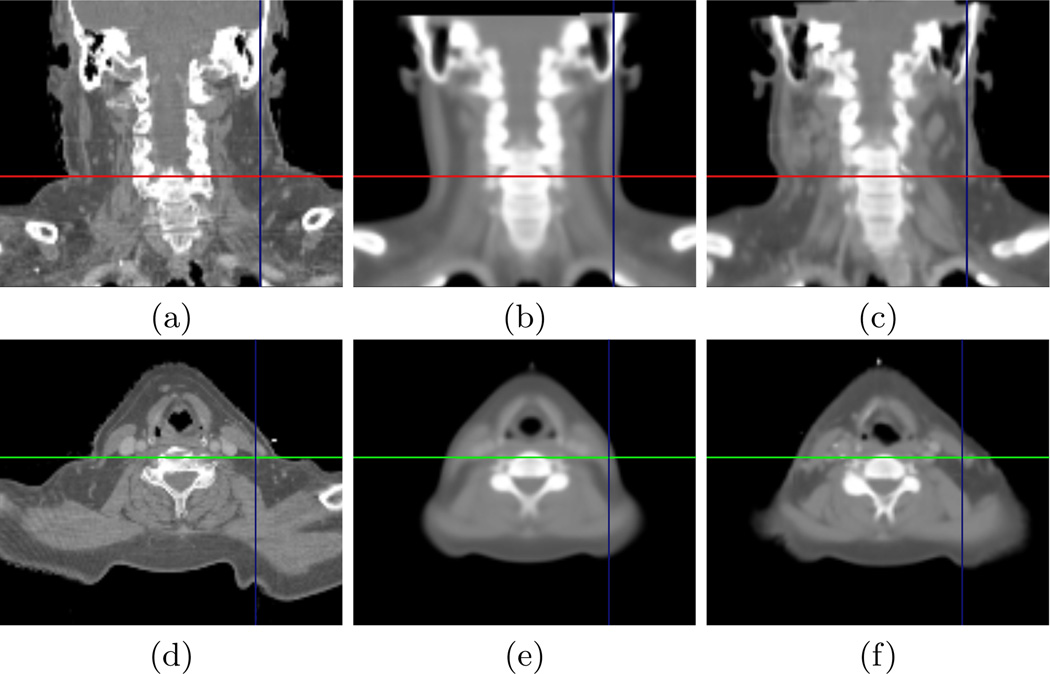
We first present in Fig. 1 the visual comparison of the average atlas and locally most similar image for one patient. This example illustrates very well that the average atlas anatomy may be significantly different from the patient after a global registration. This may result in registration discrepancies and in erroneous segmentations. This is particularly visible in the lymph nodes areas (see axes in the images) where the patient is much more corpulent than the atlas. The composite patient is much closer visually to the patient. The deformations between these two images will therefore be easier to recover and this will contribute to minimize the registration errors.
Fig. 1. Example of Computed Piecewise Most Similar Image.
Comparison between the patient image (a), (d) ; the average atlas (b), (e) and the locally most similar image (c), (f). All images are globally registered on the patient’s image.
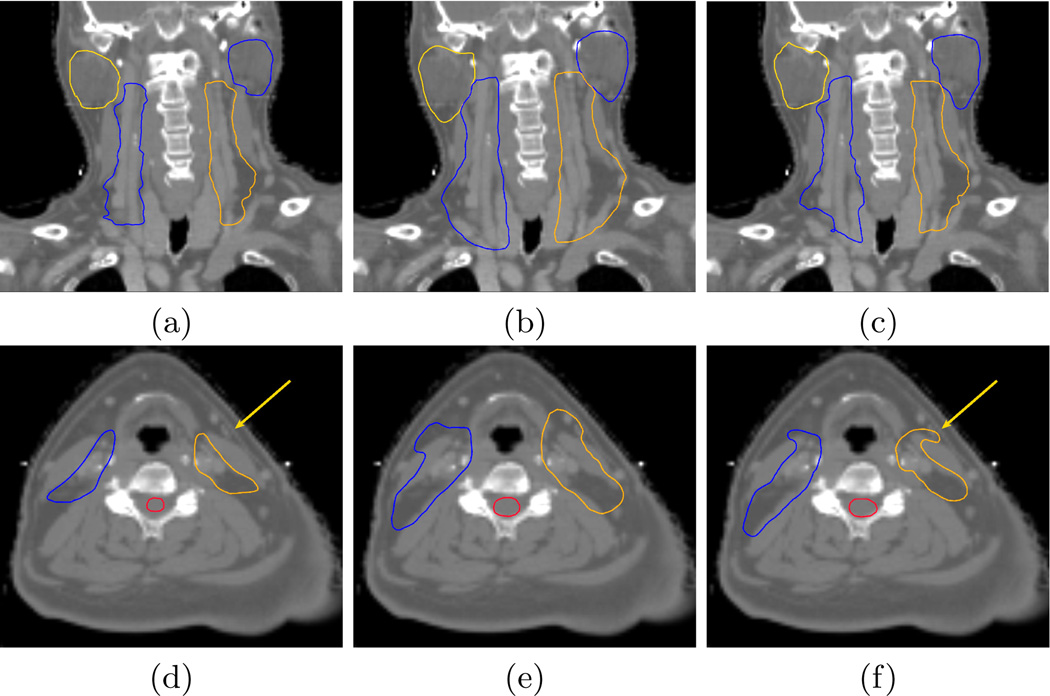
We then present in Fig. 2 the qualitative evaluation of the segmentation results, using the Leave-One-Out framework. This figure first shows that the atlas-based segmentations (images (b,e)) are overly large when compared to the manual ones. This is due to the inter-expert segmentation variabilities when creating the average segmentations, resulting in overly large segmentations in the atlas itself. This over-segmentation almost disappears using our approach. Only a single manual segmentation is indeed used for each structure, leading to more accurate segmentations, particularly on the lymph nodes areas. Finally, there are still differences in some regions (see arrow in image (f)). These differences are due to the local specificities of the selected manual segmentation, induced by the inter-expert segmentation variability.
Fig. 2. Qualitative atlas and piecewise most similar image based segmentation results.
Comparison between the manual segmentations (a), (d) ; the atlas-based segmentations (b), (e) and the piecewise most similar image segmentations (c), (f).
4.2 Quantitative Evaluation
We finally present in this section the quantitative evaluation of the results using the Leave-One-Out framework described in section 3. We present in Table 1 the average quantitative results (sensitivity and specificity) computed using the Leave-One-Out framework on 58 patients. We also indicate the number of structures on which each average was performed. The patients in the database were indeed not totally segmented manually and we therefore computed the quantitative values for the available structures.
Table 1. Quantitative Segmentation Results Comparison.
Sensitivities (Sens.), specificities (Spec.) and their standard deviations for atlas-based and locally most similar image (Frankenstein) based segmentation (see text).
| Atlas | Frankenstein | Patients | |||
|---|---|---|---|---|---|
| Sens. ± StD | Spec. ± StD | Sens. ± StD | Spec. ± StD | ||
| Lymph Nodes (L) | 0.930 ± 0.051 | 0.607 ± 0.070 | 0.692 ± 0.097 | 0.813 ± 0.072 | 53 |
| Lymph Nodes (R) | 0.923 ± 0.045 | 0.630 ± 0.078 | 0.675 ± 0.113 | 0.832 ± 0.074 | 46 |
| Spinal Cord | 0.938 ± 0.044 | 0.730 ± 0.065 | 0.773 ± 0.093 | 0.867 ± 0.079 | 47 |
| Left Parotid | 0.885 ± 0.072 | 0.691 ± 0.089 | 0.700 ± 0.172 | 0.813 ± 0.074 | 22 |
| Right Parotid | 0.879 ± 0.085 | 0.703 ± 0.078 | 0.684 ± 0.107 | 0.856 ± 0.050 | 19 |
This table shows an important improvement of the specificity measure in the locally most similar method with respect to classical atlas-based segmentation. This confirms the observations made in the qualitative results as this measure increases as the over-segmentation of the structures with respect to the manual segmentations decreases. However, the sensitivity is lower in the locally most similar case. This is mainly due to the inter-expert segmentation variabilities illustrated in Fig. 2. Nevertheless, these results are very promising and show that the locally most similar image allows to obtain an atlas whose anatomy is close to the patient to delineate and to remove the over-segmentation.
5 Conclusion
We have presented a new method to select, up to an affine transformation, the locally most similar images associated to regions of interest predefined on a precomputed average atlas. This is based on the use of a Log-Euclidean distance between transformations obtained through atlas construction and the transformation to register the patient on the atlas. As the atlas is pre-computed, the selection method is very efficient, requiring only one non linear registration. We have then associated to this selection a novel framework to build from these images a composite patient to be used as a template for the patient segmentation.
This method was validated by comparing it to atlas-based segmentation on a subset of 58 CT images among a database of 105 head and neck patients. The segmentations are not over-segmented anymore using our approach. This is seen both on qualitative and quantitative results (specificity). This method has a great interest and could also be applied to many other regions where large variabilities may be seen in the patients anatomies, such as the abdomen region.
We have seen in our experiments that the sensitivity results are corrupted by a large intra- and inter-expert segmentation variability. It is also partially responsible for the overly large segmentations in the average atlas. In the future, we aim at studying further this segmentation variability by computing locally, in the average atlas reference frame, the changes of each delineation, for example using the Staple algorithm [14]. This would be of great interest to reduce the variability inuence on the segmentation process.
Finally, we will study in the future other selection criterions, such as intensity based comparisons between the images, and compare their performance on the selection of the locally most similar image. We will also study how to combine these different criterions to obtain a very robust selection criterion.
Acknowledgments
This investigation was supported in part by a research grant from CIMIT and by NIH grants R03 CA126466, R01 RR021885, R01 GM074068 and R01 EB008015. This work was undertaken in the framework of the MAESTRO project (IP CE503564) funded by the European Commission.
References
- 1.Bondiau P, Malandain G, Chanalet S, Marcy P, Habrand J, Fauchon F, Paquis P, Courdi A, Commowick O, Rutten I, Ayache N. Atlas-based automatic segmentation of MR images: validation study on the brainstem in radiotherapy context. Int J Radiat Oncol Biol Phys. 2005;61(1):289–298. doi: 10.1016/j.ijrobp.2004.08.055. [DOI] [PubMed] [Google Scholar]
- 2.Poon I, Fischbein N, Lee N, Akazawa P, Xia P, Quivey J, Phillips T. A population-based atlas and clinical target volume for the head and neck lymph nodes. Int J Radiat Oncol Biol Phys. 2004;59(5):1301–1311. doi: 10.1016/j.ijrobp.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 3.Zhang T, Chi Y, Meldolesi E, Yan D. Automatic delineation of on-line head and neck computed tomography images: Toward on-line adaptive radiotherapy. Int J Radiat Oncol Biol Phys. 2007;68(2):522–530. doi: 10.1016/j.ijrobp.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 4.Guimond A, Meunier J, Thirion J. Average brain models: A convergence study. Computer Vision and Image Understanding. 2000;77(2):192–210. [Google Scholar]
- 5.Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage. 2004;23(Suppl 1) doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 6.Commowick O, Grégoire V, Malandain G. Atlas-based delineation of lymph node levels in head and neck computed tomography images. Radiotherapy Oncology. 2008;87(2):281–289. doi: 10.1016/j.radonc.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 7.Grégoire V, Levendag P, Ang KK, Bernier J, Braaksma M, Budach V, Chao C, Coche E, Cooper JS, Cosnard G, Eisbruch A, El-Sayed S, Emami B, Grau C, Hamoir M, Lee N, Maingon P, Muller K, Reychler H. CT-based delineation of lymph node levels and related CTVs in the node-negative neck: DA-HANCA, EORTC, GORTEC, NCIC, RTOG consensus guidelines. Radiotherapy Oncology. 2003;69(3):227–236. doi: 10.1016/j.radonc.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Blezek DJ, Miller JV. Atlas stratification. MedIA. 2007;11(5):443–457. doi: 10.1016/j.media.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aljabar P, Heckemann R, Hammers A, Hajnal JV, Rueckert D. Classifier selection strategies for label fusion using large atlas databases; Proc. of MICCAI’07, Part I. Volume 4791 of LNCS; 2007. pp. 523–531. [DOI] [PubMed] [Google Scholar]
- 10.Shelley M. Frankenstein. Lackington, Hughes, Harding, Mavor and Jones; 1818. [Google Scholar]
- 11.Arsigny V, Commowick O, Pennec X, Ayache N. MICCAI (I). LNCS. 4190. 2006. A Log-Euclidean framework for statistics on diffeomorphisms; pp. 924–931. [DOI] [PubMed] [Google Scholar]
- 12.Pitiot A, Bardinet E, Thompson PM, Malandain G. Piecewise affine registration of biological images for volume reconstruction. MedIA. 2006;10(3):465–483. doi: 10.1016/j.media.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Arsigny V. Processing Data in Lie Groups: An Algebraic Approach. Application to Non-Linear Registration and Diffusion Tensor MRI. PhD, Polytechnique. 2006 [Google Scholar]
- 14.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE TMI. 2004;23(7):903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]