Figure 6. Tdg-depletion induced 5fC/5caC signals are enriched at bivalent and transcriptionally silent gene promoters in mouse ESCs.
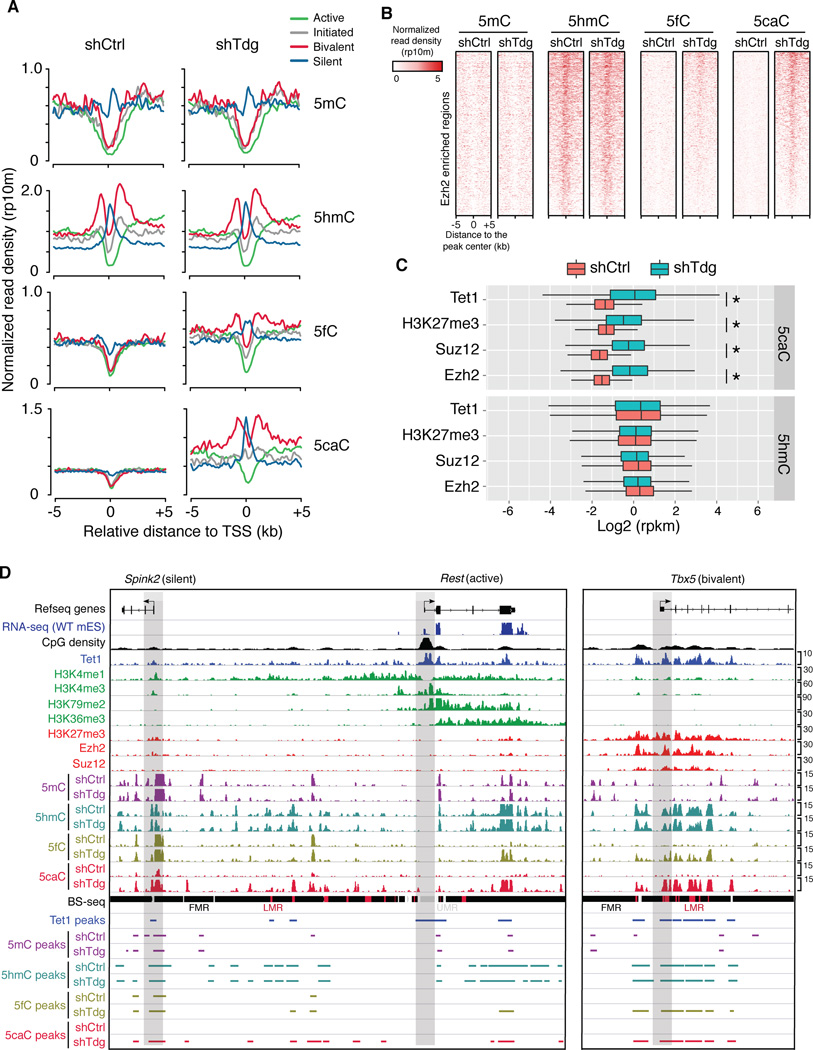
(A) Average 5mC/5hmC/5fC/5caC signals in control and Tdg-deficient mouse ESCs at TSSs of four groups of gene promoters that are associated with distinct chromatin states (active: H3K4me3+/H3K79me2+; initiated: H3K4me3+ only; bivalent: H3K4me3+/H3K27me3+; silent: none).
(B) 5mC/5hmC/5fC/5caC levels in control and Tdg-deficient mouse ESCs at representative loci of gene promoters that are associated with different histone modification patterns. The gene promoters are highlighted by grey bars. Fully methylated regions (FMRs), low methylated regions (LMRs) and unmethylated regions (UMRs) were derived from previously published BS-seq datasets.
(C) Heat maps of 5mC/5hmC/5fC/5caC levels (normalized read density) in control and Tdg-deficient mouse ESCs at centers of Ezh2 binding sites.
(D) Box-plots of normalized 5hmC and 5caC levels (read per million reads and kilo bases, rpkm) in control and Tdg-deficient cells within genomic regions enriched for Tet1, Ezh2, Suz12 and H3K27me3. *, P<2.2e-16 (P-values were calculated by two-tailed t-test).
See also Figure S6.