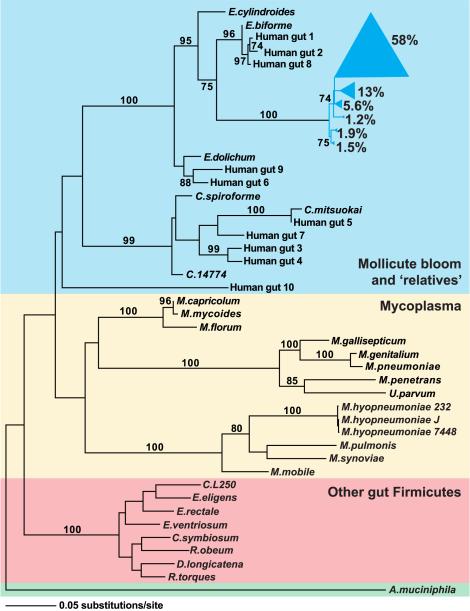
Figure 4. Phylogeny of selected representatives from the Firmicutes division, including the Mollicute bloom and closely related human strains.
16S rRNA gene sequences for previously sequenced Firmicute genomes and Mollicute strains isolated from the human gut were identified in the RDP database (Cole et al., 2005). All Mollicute sequences obtained from conventionalized C57BL/6J mice fed a CHO or Western diet (n=801 sequences) and from our previous survey of obese humans (length>1250 nucleotides; n=571 sequences; Ley et al., 2006b) were separately binned into phylotypes using DOTUR (99% identity; Schloss and Handlesman, 2005). One representative of each of the six dominant mouse phylotypes was chosen (together comprising 81% of the mouse Mollicute sequences) in addition to one representative of each of the ten dominant human phylotypes. Likelihood parameters were determined using Modeltest (Posada and Crandall, 1998) and a maximum-likelihood tree was generated using PAUP (Swofford, 2003). Bootstrap values represent nodes found in >70 of 100 repetitions. Phylotypes from the Mollicute bloom are shown in blue; wedge size is proportional to the indicated relative abundance (% of Mollicute 16S rRNA gene sequences). The Mollicute bloom and relatives are shaded in blue, previously sequenced Mollicutes (including the obligate parasites, Mycoplasma, and Mesoplasma florum) are shaded in yellow, and recently sequenced Firmicutes found in the ‘normal’ distal human gut microbiota are shaded in red. Akkermansia muciniphila, a Verrucomicrobia, was used to root the tree (shaded in green).