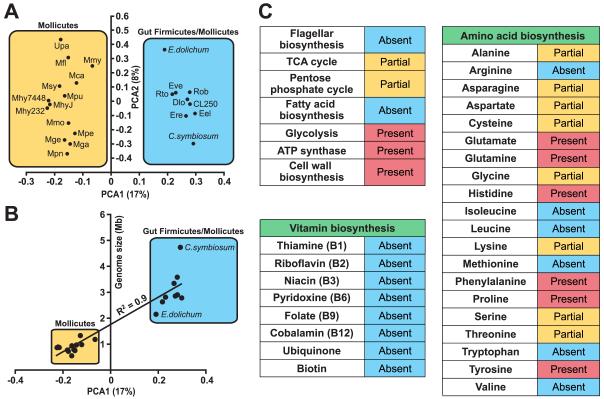
Figure 6. Metabolic and principal component analysis (PCA) of sequenced Firmicute genomes.
(A) PCA analysis of 14 previously sequenced Mollicute genomes (mostly Mycoplasma) and draft genome assemblies of nine human gut-associated Firmicutes (http://genome.wustl.edu/pub/). MetaGene was used to predict proteins from each genome (Noguchi et al., 2006). Proteins were then assigned to KEGG orthologous groups based on homology (BLASTP e-value<10−5; KEGG version 40; Kanehisa et al., 2004). Genomes were clustered based on the relative abundance of KEGG metabolic pathways (number of assignments to a given pathway divided by total number of pathway assignments). Only pathways found at >0.6% relative abundance in at least two genomes were included. The first two components are shown, representing 17% and 8% of the variance respectively. Abbreviations: Mca, Mycoplasma capricolum; Mfl, Mesoplasma florum L1; Mga, Mycoplasma gallisepticum R, Mge, Mycoplasma genitalium G37; Mhy232, Mycoplasma hyopneumoniae 232; Mhy7448, Mycoplasma hyopneumoniae 7448; MhyJ, Mycoplasma hyopneumoniae J; Mmo, Mycoplasma mobile 163K; Mmy, Mycoplasma mycoides subsp. mycoides SC str. PG1; Mpe, Mycoplasma penetrans HF-2; Mpn, Mycoplasma pneumoniae M129; Mpu, Mycoplasma pulmonis UAB CTIP; Msy, Mycoplasma synoviae 53; Upa, Ureaplasma parvum; E.dolichum, Eubacterium dolichum; CL250, Clostridium sp. L2-50; C.symbiosum, Clostridium symbiosum; Dlo, Dorea longicatena; Eel, Eubacterium eligens; Ere, Eubacterium rectale; Eve, Eubacterium ventriosum; Rob, Ruminococcus obeum; and Rto, Ruminococcus torques. (B) KEGG pathway relative abundance has a significant correlation with genome size. A linear regression was performed comparing PCA1 to genome size (or draft assembly size). PCA1 has a significant correlation to genome size (R2=0.9, p<0.05). (C) Metabolic pathways in E.dolichum. Pathways are marked partial if most genes are present and absent if ≤2 genes are present.