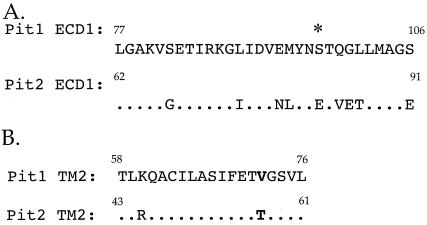
FIG. 5.
Amino acid sequence comparison of putative ECD1 and TM2 of PiT1 and PiT2. (A) Replacement of PiT1 ECD1 with that of PiT2 renders PiT1 functional as an A-MuLV receptor. The amino acid sequence identity between ECD1 of PiT1 and PiT2 is approximately 70% and includes conservation of the N-linked glycosylation site (*). (B) Alignment of PiT1 and PiT2 TM2 reveals approximately 89% residue identity between the two domains. The bold text represents the difference between PiT1 (valine) and PiT2 (threonine) at PiT1 position 72, which is thought to play a role in determining receptor topology. Numbers indicate where the respective domains begin and end.