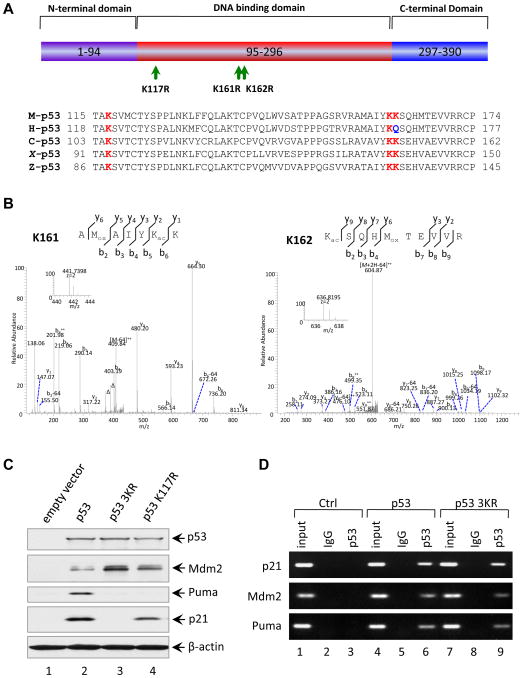
Figure 3. Loss of Acetylation Sites at K117, K161 and K162 of Mouse p53 Abolishes the Activation of p21 and PUMA.
(A) Schematic representation of the mouse p53 protein with three mutated acetylation sites indicated and alignment of the K117, K161 and K162 flanking regions of the mouse p53 with those of other species. The conserved lysines were highlighted in red and Q165 of human p53 highlighted in blue is evolutionarily reminiscent of acetylated K.
(B) Mass spectrometry analysis of tryptic mouse p53 peptides containing K161 and K162. The fragmentation spectrum of 156AMoxAIYKacK162 and 162 KacSQHMoxTEVVR171 revealed the presence of peptides with acetylation at K161 and K162, respectively. The protein was prepared as described in the Experimental Procedures. Inset shows the high-resolution precursor ion mass. The label “Δ” designates “b” or “y” ions with water and/or ammonia loss. “Kac” and “Mox” designate acetyllysine and oxidized methionine, respectively. Neutral loss of sulfenic acid from oxidized methionine was indicated as “-64”.
(C) Western blot analysis of p53, Mdm2, Puma and p21 in H1299 cells transfected with plasmids expressing mouse wildtype p53, p53-K117R and p53-3KR mutants. β-actin was used as a loading control.
(D) ChIP assay for the binding of p53 or p53-3KR mutant to the consensus sites in the p21, Puma and Mdm2 promoters. H1299 cells transfected with plasmids DNA expressing mouse p53 wildtype (WT) and p53-3KR mutant were treated with 1% formaldehyde for 10 min and processed for ChIP analysis. The occupancy of p53 or p53-3KR of the p21, Puma and Mdm2 promoters were detected by PCR-agarose gel electrophoresis. IgG antibody serves as a negative control.