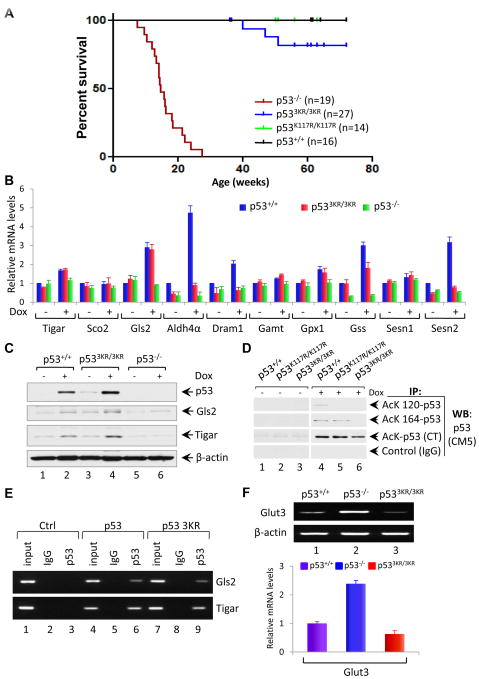
Figure 6. p533KR Retains Its Tumor Suppressor Activity and the Ability to Activate Metabolic Targets.
(A) Kaplan-Meier survival curves of p53+/+, p53K117R/K117R, p533KR/3KR and p53−/− mice.
(B) qRT-PCR analysis of indicated mRNAs in p53+/+, p533KR/3KR and p53−/− MEFs either untreated or treated with 0.2 μg/ml Dox for 8 hours. Data are averages ± SEM of each mRNA quantities normalized first to β-actin and then to the untreated wildtype sample values from three independent MEF lines.
(C) Immunoblot assays of p53, Gls2 and Tigar protein in the whole cell lysates prepared from p53+/+, p533KR/3KR and p53−/− MEFs either untreated or treated with 0.2 μg/ml Dox for 8 hours. β-actin serves as a loading control.
(D) Analysis of acetylation of p53 at K117, K161 and K162 in p53K117R/K117R and p533KR/3KR MEF cells. p53+/+ and p533KR/3KR and p53K117R/K117R MEFs either untreated or treated with 0.2 μg/ml Dox, 1.0 μM TSA and 5 mM Nicotinamide for 6 hours. Cell lysates were immunoprecipitated with either anti-AcK120-p53, anti-AcK164-p53, anti- AcK CT-p53 antibodies or control IgG, and blotted with anti-p53 (CM5) antibody using ReliaBLOT systems from Bethyl Laboratories.
(E) ChIP assay for the binding of p53 or p53-3KR mutant to the consensus sites of p53 metabolic targets Gls2 and Tigar in H1299 cells transfected with plasmids DNA expressing mouse p53 wildtype (WT) or p53-3KR mutant.
(F) RT-PCR (upper panel) and qRT-PCR (lower panel) analysis of Glut3 expression in p53+/+, p533KR/3KR and p53−/− primary MEFs at passage 2. Data are represented as averages ± SEM from three independent experiments.
See also Figure S7