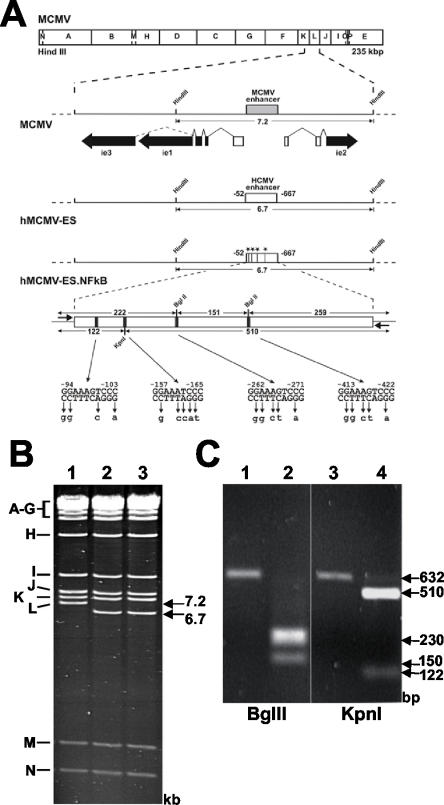
FIG. 1.
Mutagenesis of the HCMV-MIEP NF-κB binding sites in hMCMV-ES. (A) Organization of the MIE regions of the constructed MCMV BAC genomes. The HindIII map of the MCMV genome is given at the top. The expanded map below represents the MIE region of MCMV with the enhancer (gray box) and the exon-intron structure of the ie1, ie2, and ie3 genes indicated. Coding and noncoding exons are depicted as black and white boxes, respectively. The BAC plasmids hMCMV-ES and hMCMV-ES.NF-κB carry a 616-bp fragment (white rectangle) from the HCMV MIE promoter in replacement of the MCMV enhancer. In hMCMV-ES.NF-κB, the NF-κB binding sites were mutated, introducing unique restriction sites (−98 to −103 StuI, −161 to −166 KpnI, and −265 to −270 and −416 to −421 BglII), which are indicated by asterisks or black bars. The sizes of the expected HindIII L fragments of the MCMV BAC genomes and the sizes of the expected BglII and KpnI fragments of the PCR-amplified HCMV enhancer (with primers shown with short arrows flanking the enhancer) are indicated by the arrows. Coordinates are given relative to the MCMV ie1/ie2 transcription start site. The diagrams are not drawn to scale. (B) Ethidium bromide-stained agarose gel of HindIII-digested MCMV BAC plasmid (1), hMCMV-ES (2), and hMCMV-ES.NF-κB (3). The names and sizes of the HindIII fragments of the MCMV genome are indicated in the margins. (C) To verify successful mutagenesis of NF-κB sites, enhancer sequences were amplified from MCMV-ES (lanes 1 and 3), and hMCMV-ES.NF-κB (lanes 2 and 4)BACs by PCR. The amplified products were digested with restriction enzyme BglII (lanes 1 and 2) or KpnI (lanes 3 and 4), and the resulting DNA fragments were separated by gel electrophoresis.