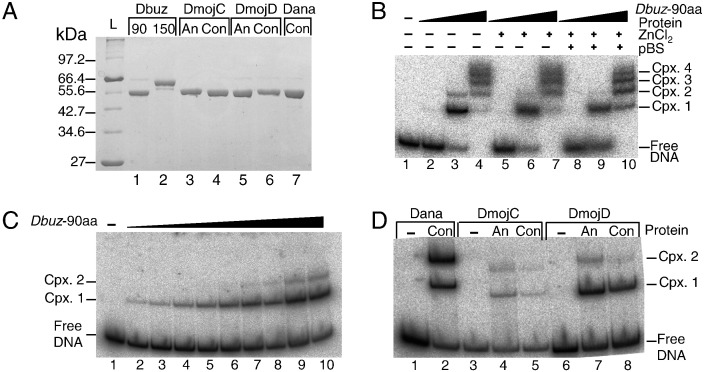
Fig. 3.
Protein expression, purification and DNA binding.
A) SDS-PAGE with the 7 expressed THAP domain proteins, ~ 5 μg of each protein was loaded as indicated. D. buzzatii protein was either 90 or 150 amino acids from the N-terminus of the transposase. The others were 90 amino acids long.
B) EMSA performed with Dbuz\Galileo-THAP-90aa. Three different binding conditions were tested. First lane is Dbuz\GalileoG labeled TIR (2.2 nM). Lanes 2, 3 and 4 are × 100 increasing protein concentrations (470 pM, 47 nM and 4.7 μM). Lanes 5, 6 and 7 are the same but with 100 μM ZnCl2. Lanes 8, 9 and 10 are the same but with 500 ng of pBluescript as competitor. Note that the proteins were purified in a buffer containing EDTA and reactions in which zinc was not added back contained only that zinc acquired by the proteins during folding.
C) Fine titration EMSA of the Dbuz\Galileo-THAP-90aa with its TIR (0.14 nM). Protein concentration increases 2-fold in successive lanes: 0.184 nM, 0.367 nM, 0.734 nM, 1.469 nM, 2.938 nM, 5.875 nM, 11.75 nM, 23.5 nM, 47 nM and 94 nM.
D) EMSA in which binding of the indicated 90 amino acid THAP domains is tested against the consensus TIR of their respective Galileo sub-group. Final protein concentration: 5.8 nM and TIR final concentration is 0.28 nM. Note that it is not necessary to reconstruct the ancestral TIR because, unlike a transposase gene, a non-functional TIR can not be amplified by transposition. The consensus TIR can therefore be expected to retain functionality.