Fig. 6.
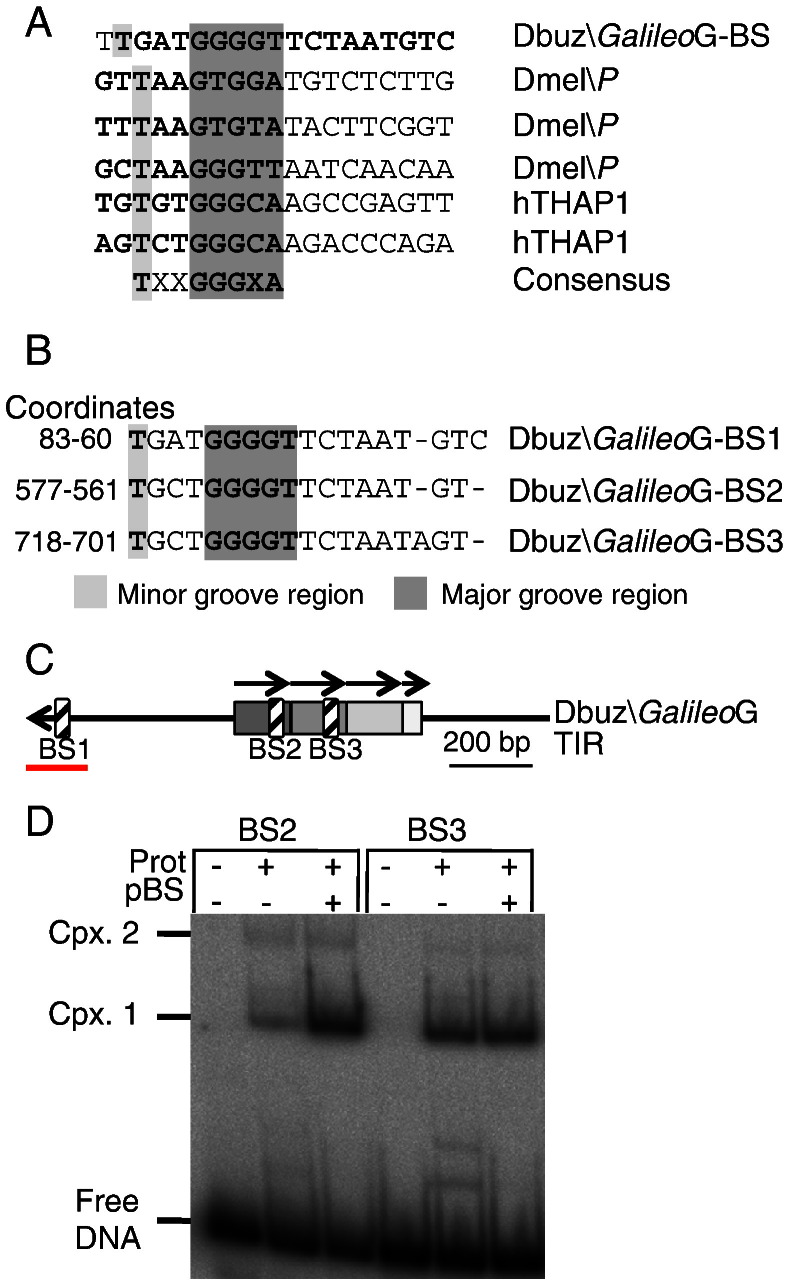
THAP domain binding sequence comparison.
A) Dbuz\GalileoG compared to Dmel\P-element and hTHAP1 binding sites (Bessière et al., 2008; Campagne et al., 2010; Sabogal et al., 2010). The major and minor groove interacting regions are colored. A putative consensus THAP binding sequences, including Dbuz\GalileoG sequences is deduced.
B) Alignment of the Dbuz\GalileoG binding site with other putative binding sites found downstream in the Dbuz\GalileoG-TIR.
C) Structure of the Dbuz\GalileoG-TIR where the tandem repeats are drawn as grey rectangles and the binding sites are drawn with hatched shading (BS1, BS2 and BS3). The red bar depicts the 150 bp TIR consensus region used in the different experiments. A 200 bp scale bar is also provided.
D) The putative secondary binding sites in Galileo are functional. 50 bp oligonucleotides encoding the putative Galileo secondary binding sites identified in part C were tested for binding in an EMSA using the Dbuz\GalileoG-90aa DNA binding domain (47 nM). pBluescript (500 ng) was added as an non-specific competitor. The location of BS2 and BS3 is shown in part C.