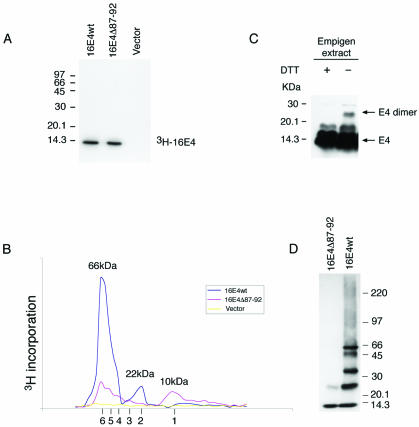
FIG. 6.
16E1∧E4 self-associates to form multimers of 66 kDa. (A) The 3H-labeled 16E1∧E4 protein generated by in vitro transcription-translation was immunoprecipitated using anti-16E1∧E4 rabbit polyclonal antibody and separated by SDS-PAGE. Both wt and mutant (Δ87-92) E1∧E4 proteins gave a single band of ∼10 kDa. Molecular mass standards are shown on the left. (B) The 3H-labeled 16E1∧E4 proteins shown in panel A were separated on a Superdex 75 column, and the elution profile was obtained following scintillation counting. The column was calibrated using globular standards, and the molecular masses of the major 16E1∧E4 peaks are shown above the profiles. wt 16E1∧E4 eluted as a major peak of 66 kDa, which corresponds to the predicted size of 16E1∧E4 hexamers. A minor peak of 22 kDa was also apparent. Mutant 16E1∧E4 (Δ87-92) eluted as peaks of increasing molecular mass, with the smallest peak corresponding to the size of 16E1∧E4 monomers (10 kDa). The expected elution positions of E1∧E4 multimers (monomers to hexamers) are shown beneath the profile. (C) Empigen extracts from 16E1∧E4-expressing SiHa cells were analyzed by SDS-PAGE and Western blotted following treatment with DTT. The positions of monomeric 16 E1∧E4 and disulfide-stabilized 16E1∧E4 dimers are shown on the right. Complexes larger than dimers were not apparent following extraction using Empigen, which is necessary to solubilize the cytoskeleton-associated E1∧E4 protein. Molecular mass standards are shown on the left. +, present; −, absent. (D) wt 16E1∧E4 protein purified from E. coli migrates as a series of bands of increasing molecular mass when analyzed by gel electrophoresis in the presence of low levels of SDS. The largest band has a molecular mass of 66 kDa, in agreement with the results obtained using fast protein liquid chromatography. The C-terminal truncation (16E1∧E4Δ87-92) exists only as monomers and dimers.