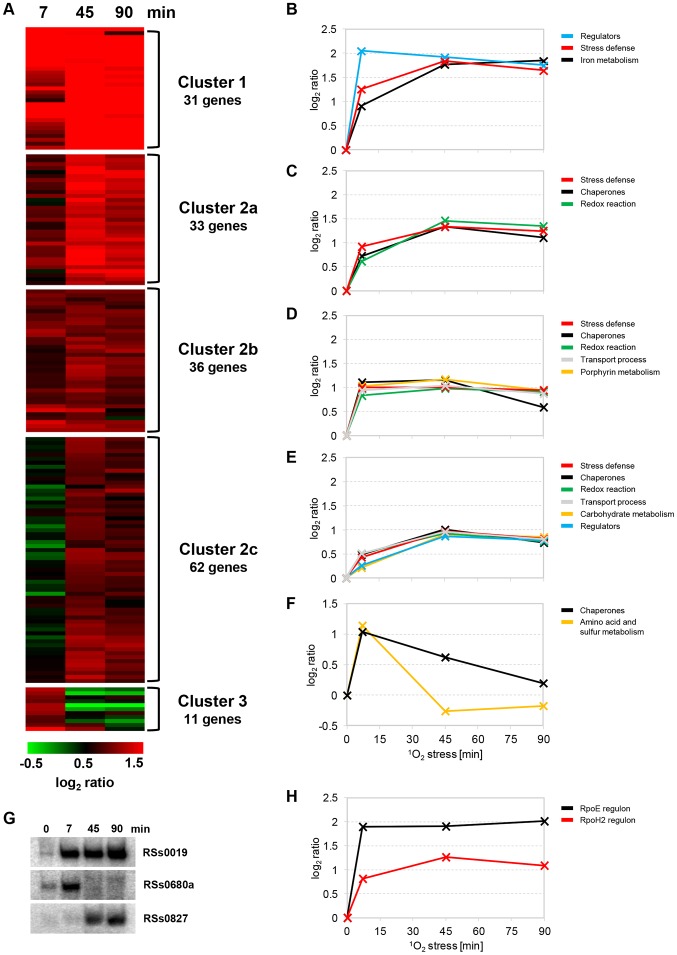
Figure 2. Expression kinetics of stress-related mRNAs and sRNAs.
Relative changes of mRNAs upon singlet oxygen (1O2) stress were monitored by microarray analysis of total RNA at time-points 7, 45, and 90 min and depicted as log2 ratios. (A) All mRNAs with significant induction (log2 ratio ≥0.8 and p-value <0.05) at one of the three time-points were applied to clustering using MeV (Multi Experiment Viewer version 4.7.4) from the TM4 Microarray Software Suite [67], [68]. Clustering was based on k-means (KMC method) according to Euclidean distance with a maximum of 50 iterations. Changes were illustrated as heat-maps with a color code ranging from −0.5 (green) to 1.5 (red) log2 ratio. Columns represent time-points of the stress response and rows represent individual genes. (B–F) Up-regulated mRNAs shown in (A) were grouped together according to their function. Expression kinetics, representing the mean of log2 ratios, were calculated for functional groups of cluster 1 (B), cluster 2a (C), cluster 2b (D), cluster 2c (E), and cluster 3 (F). (G) Analysis of stress-induced sRNAs. Total RNA was isolated at the indicated time-points and applied to Northern blot analysis. RSs0019, RSs0680a, and RSs0827 were hybridized to radioactively labeled oligonucleotides and visualized by phosphoimaging. 5S rRNA was probed as a control for equal loading of samples (not shown). (H) Genes with described RpoE- and RpoH2-dependent promoters [28], [29], [72] were analyzed when they exhibited log2 ratios ≥0.8. The curves represent the mean of log2 ratios based on 12 mRNAs (RpoE, black line) and 42 mRNAs (RpoH2, red line). See supplementary Tables S1 and S2 for further information on regulated genes and their particular functions.