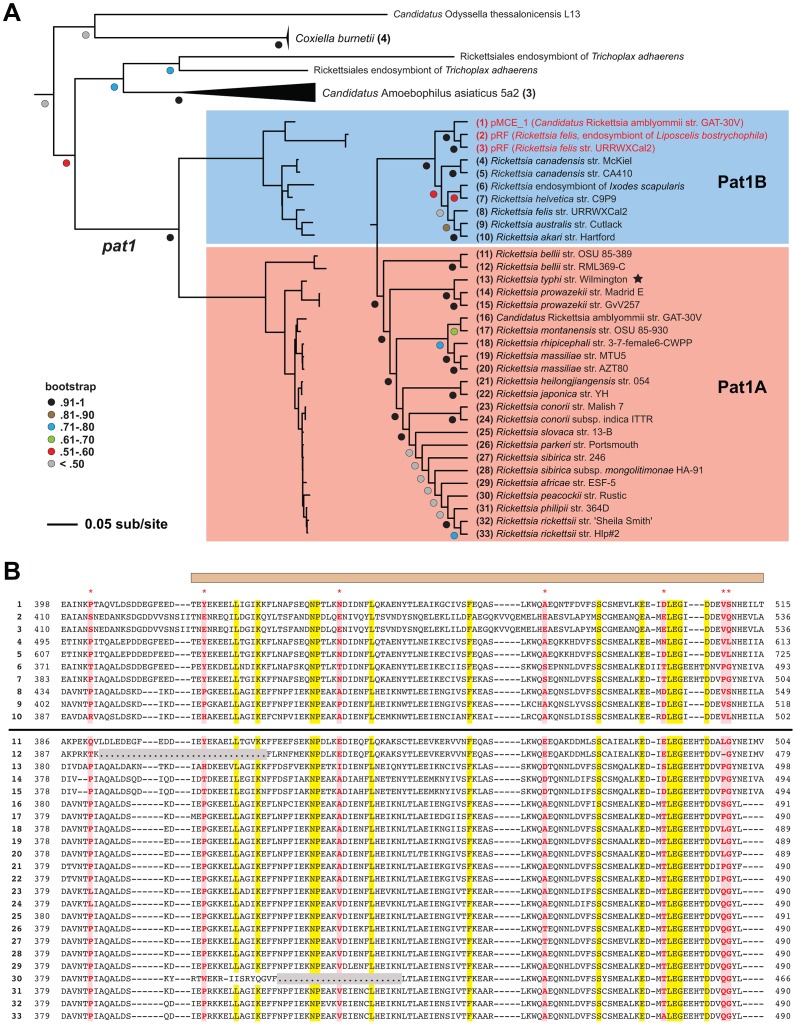
Figure 2. Evolutionary analysis of rickettsial Pat1 proteins.
A) Phylogeny estimation of rickettsial Pat1 proteins, with closest non-Rickettsia clades shown. The complete tree containing 93 ExoU-like patatins is shown in Fig S2. See Materials and Methods for details of phylogenetic analysis. The Rickettsia Pat1 sequences (n = 33) form two well-supported clades (Pat1A and Pat1B). The Pat1B group (1–10, shaded blue) contains the plasmid-encoded Pat1 proteins (colored red), Pat1 of the R. canadensis strains, and the proteins from REIS, R. helvetica (incertae sedis) and the transitional group rickettsiae. The Pat1A group (11–33, shaded blue) contains R. typhi RT0590 (noted with a black star), the R. prowazekii and R. bellii proteins, and Pat1 from the majority of the spotted fever group rickettsiae. B) Comparative analysis of the C-terminal tails for 33 rickettsial Pat1 proteins. See text for alignment details. The sequences are listed (1–33) as they appear in the phylogenetic tree in panel A. The horizontal bar distinguishes Pat1B (upper) from Pat1A (lower) sequences. The tan bar above the alignment illustrates the delineation between the tail region and the variable region of Pat1 sequences. Coordinates for each sequence are provided at left and right. Conserved positions across the alignment are highlighted yellow, with positions predicted to evolve under positive selection [71] highlighted red and noted at the top of the alignment by asterisks. Positions depicted with dots and highlighted gray illustrate missing sequence for Pat1 proteins encoded by two ORFs (R. bellii str. RML369-C and R. peacockii str. Rustic).