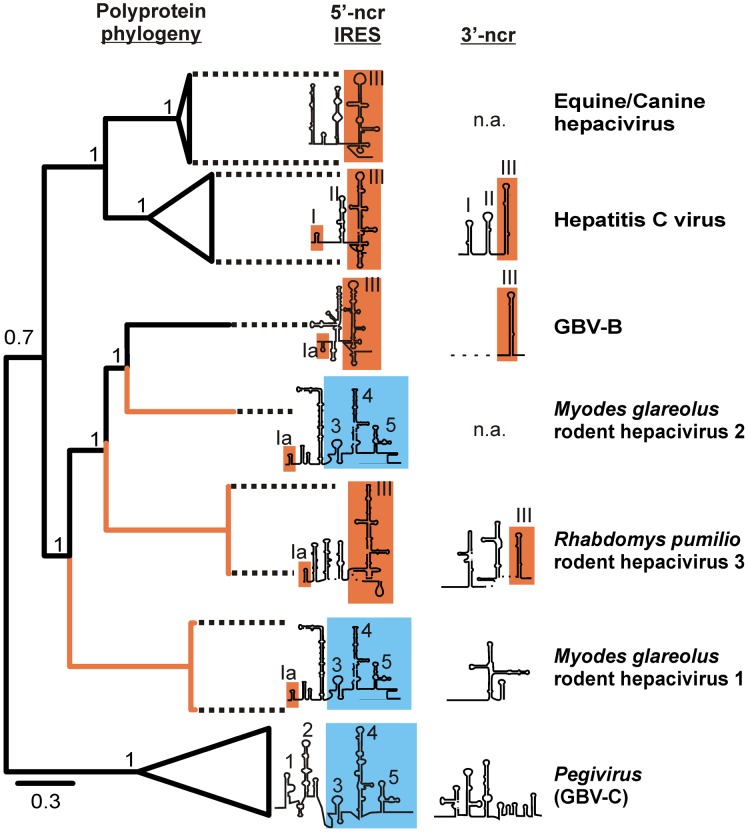
Figure 5. Complete polyprotein gene phylogeny and comparison of the genome termini between the novel rodent and prototype hepaciviruses.
For the Bayesian phylogeny shown to the left, the WAG amino acid substitution model was used in MrBayes as indicated in the methods section. Statistical support of grouping from Bayesian posterior probabilities is indicated at node points. Scale bar corresponds to genetic distance. The Pestivirus BVDV (NC_001461) was chosen as an outgroup and truncated for graphical reasons. Branches leading to the novel hepaciviruses from this study are in orange. GenBank accession numbers of analyzed hepaciviruses correspond to those indicated in Figure 3A . The 5′- and 3′-genome termini were re-drawn from published foldings for equine hepaciviruses [24], HCV [63], [77] and GBV-B [78] and de novo for this study for the 5′- and 3′-ends of GBV-C1 and the 3′-end of GBV-B (see Supplementary Figure 4). Despite earlier attempts to fold the 3′-ncr of GBV-B [21] only the 3′-terminal stem-loop structure of GBV-B could be reliably folded due to the single sequence available. The folding of the 3′-end of RMU10-3382 remained tentative for the same reason. No sequence information was available for the 3′-ends of the canine/equine hepacivirus clade and M. glareolus hepacivirus clade 2 (indicated as “n.a.”). Typical pegivirus domains are highlighted in blue and ordered by arabian numbers. Typical hepacivirus domains are highlighted in orange and numbered by roman numbers.