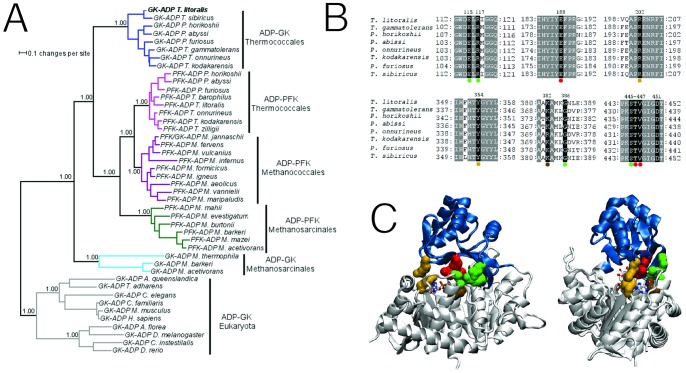
Figure 8. Conservation of cluster residues involved in domain opening/closure across the ADP-dependent sugar kinases family.
(A) Consensus phylogenetic tree for ADP-dependent sugar kinase family determined by Bayesian inference. Groups of enzymes from archaea are shown in color: GK from Thermococcales (blue), PFK from Thermococcales (pink), PFK from Methanococcales (purple) PFK from Methanosarcinales (dark green) and GK from Methanosarcinales (cyan). The group of enzymes from eukaryotic organisms that were used as an outgroup to establish the tree root is shown in gray. The posterior probability of some interesting groups is shown in its respective node. (B) Multiple sequence alignment of glucokinases from the Thermococcales group. Residues involved in clusters described in the texts are indicated by dots; cluster 1, red (Glu188-Thr446/Val447); cluster 2, yellow (Arg202-Tyr354); cluster 3, green (Arg117/Glu115-Gly386/Ser445) and brown (Lys382-Arg117). (C) Amino acids involved in clusters 1–3 are shown as spheres and colored as in B.