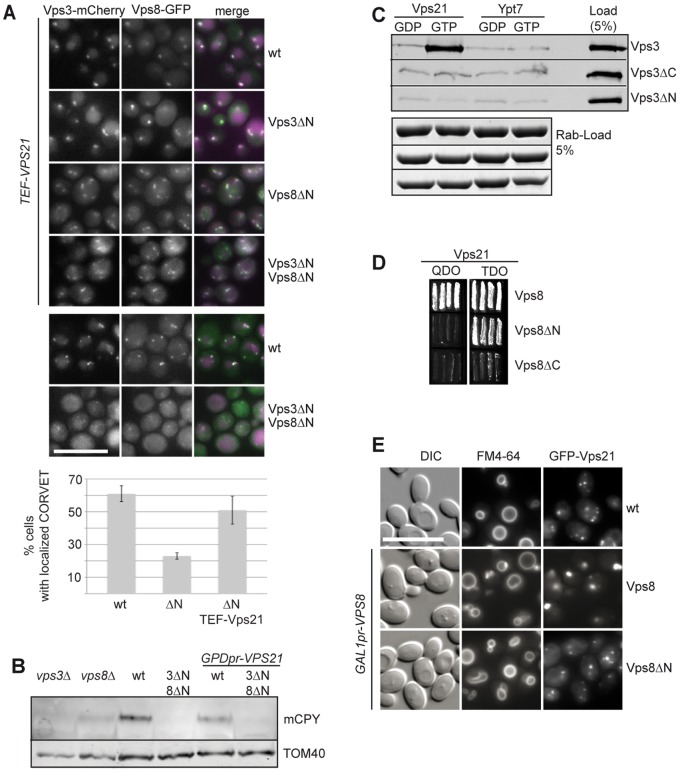
Figure 4. Connection of N-terminal domains of CORVET-specific subunits to the Rab Vps21.
(A) Localization of ΔNTD-CORVET upon overproduction of Vps21. TEF1pr-VPS21 was introduced via integrative pRS406 based plasmids into the indicated background-strains expressing the fluorescently tagged CORVET versions. Cells were monitored by fluorescence microscopy. Statistical analysis of cells with dot-localized CORVET was done by evaluating 400 cells per indicated strain. Cells with multiple dots (at least two) of colocalized Vps3 and Vps8 were counted and given by percentages. Error-bars represent standard deviation of different frames of 50 cells each. Size bar, 10 µm. (B) Functional analysis of Vps21-overexpression in truncated CORVET strains. CPY secretion assay was performed as before (Figure 1D) by isolating membrane fractions, followed by SDS-PAGE, western blot and decorating them against CPY, and TOM40 as loading-control. Vps21 was overproduced by genomically replacing the endogenous Vps21-promoter with the strong GPD3-promoter, which is even stronger than the TEF promoter used for our localization assays [26]. (C) Interaction of Vps3 with Vps21. 35 µg of yeast-purified (TAP-purification) Vps3 construct was applied to each pull-down reaction of GSH-beads coupled to GST-Vps21 or GST-Ypt7, which were loaded with GTPγs or GDP beforehand. Eluates were loaded onto SDS-PAGE, followed by western blot, which was decorated against Cbp. SDS-sample buffer was added to beads with bound Rabs afterwards, and proteins were analyzed by SDS-PAGE and Coomassie staining. For details see Methods. (D) Interaction of Vps8 with Vps21. Yeast-2-Hybrid analysis of Vps21-interaction with Vps8 constructs as preys was performed as described in Methods. Growth on QDO (quadruple drop out) plates indicates strong interactions, growth on TDO (triple drop out) refers to weaker interactions [28]. (E) Microscopy images of FM4–64 stained cells overproducing the indicated Vps8 constructs. GFP-Vps21 was used as a fluorescent marker of late endosomal accumulations caused by Vps8 overproduction. Size bar, 10 µm.