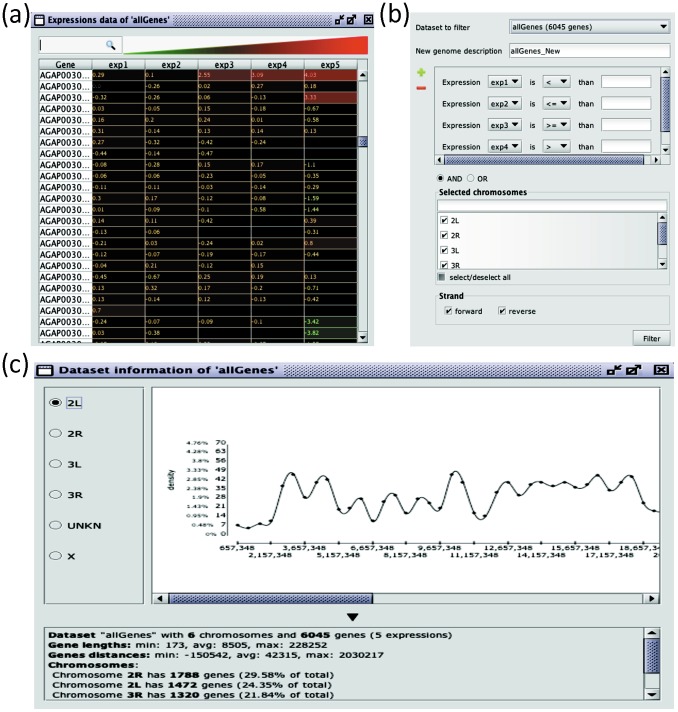
Figure 1. Expression-based gene dataset processing and Position-based gene dataset processing.
(a). An example visualization of genes with multiple expressions is shown. Expression levels are depicted by the means of different colours (black = 0, gray = not available, from green to red = increased expression values). Data are referring to transcription regulation of sex-biased genes during ontogeny in the malaria vector Anopheles gambiae [74]. The 5 different expression datasets are referring to (1st, 2nd, 3rd larvae stages, pupae and adults). (b). An example interactive dialog is shown for gene filtering based on expression level thresholding and Boolean combination of expressions. (c). An example interactive window is shown where a chromosome is rendered along with the local gene density plot. Different functions are available for retrieving gene position in the chromosome and their lengths; for retrieving the density of genes along each chromosome, for computing what percentage of a chromosome is represented by a gene dataset; for computing the min, max and average lengths of the genes populating a chromosome; the min, max and average distance between consecutive genes in a chromosome, etc. The dataset considered in this example is the Anopheles Gambiae genome.