Figure 3.
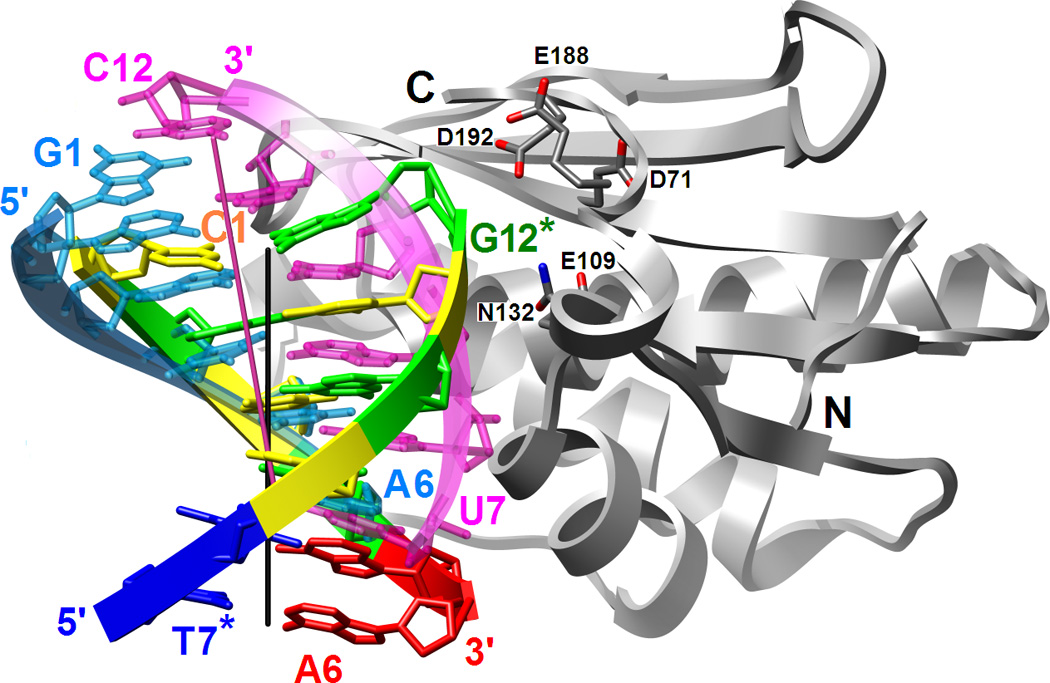

Binding mode of RNase H in the minor groove of the DDD G-tract and comparison between the relative orientations of the RNA/DNA and DNA duplexes bound to the enzyme. (A) Superimposition of the Bh-RNase HC molecules in the complexes with DDD and RNA/DNA hybrid (PDB ID code 1zbi18) reveals a two base-pair shift of the duplexes relative to one another. The enzyme adopts virtually identical conformations in the two structures and only one of the superimposed RNase H molecules is depicted. Only six base pairs per duplex are shown; the color code of residues in the DDD is the same as in Fig. 1 and DNA and RNA strands in the hybrid are colored cyan and pink, respectively. Chain termini and active site Asp and Glu(Gln) residues as well as 5′- and 3′-terminal nucleotides are labeled and solid lines in black and pink represent the overall helical axes for dsDNA and hybrid hexamers, respectively. The view is across the major and minor grooves. (B) The superimposed complexes rotated by 90° around the vertical relative to panel A and viewed into the major grooves of hexameric duplexes.
