Abstract
Chemokine are small, inducible pro-inflammatory cytokines involved in many biological processes, such as migration of leukocytes, atherosclerosis, angiogenesis, tumor growth, and metastasis. Chemokine are also known to influence tumor cell’s activity. Specifically, tumor cells express chemokine receptors in a non random manner suggesting a role of chemokine in metastatic destination of tumor cells. The present study was conducted to determine distribution of (Chemokine receptor 2) CCR2 V64I, Chemokine ligand 2 CCL2 I/D, and CCL2 2518 A>G gene polymorphisms in North Indian population and compare with different populations globally. Polymerase chain reaction (PCR)-based analysis was conducted in 200 normal healthy individuals of similar ethnicity. Allelic frequencies in wild type (GG) of CCR2 V64I G>A were 63 % G; CCL2 I/D 42 % II; CCL2 2518 A>G 40.5 % A. The minor variant allele frequency in our population was as follows: 19.5 % for CCR2 V64I, 35.5 % for CCL2 I/D, 35.3 % for CCL2 2518 A>G. We further compared frequency distribution for these genes with various published studies in different ethnicity. Our results suggested that frequency in chemokine genes exhibit distinctive pattern in India that could be attributed to ethnicity variation. This could assist in high-risk screening of human exposed to environmental carcinogens and cancer predisposition in different ethnic groups. Thus, they signify an impact of ethnicity and provide a basis for future epidemiological and clinical studies.
Keywords: Chemokine gene, SNPs, Bladder cancer, PCR–RFLP, North India
Introduction
Genetic variation plays a critical but largely uncharacterized role in differentiation. Variations in the DNA sequences of humans can affect how they develop and respond to pathogens, chemicals, drugs, vaccines, and other agents. Genetic variation in human genome is an emerging resource for studying cancer, a complex disease characterized by both environmental and genetic contributions. Inflammation is a multistep process that includes injury, repair, and resolution. Chronic inflammation can incite carcinogenesis by inducing proliferative events and post-translational DNA modifications by enhancing the secretion of growth factors such as cytokines and chemokines, and inducing oxidative stress by the release of nitric oxide (NO) and reactive oxygen species (ROS). Chemokines are secreted, low molecular weight proteins (8–30 kDa) that bind to seven transmembrane protein-coupled receptors [1].
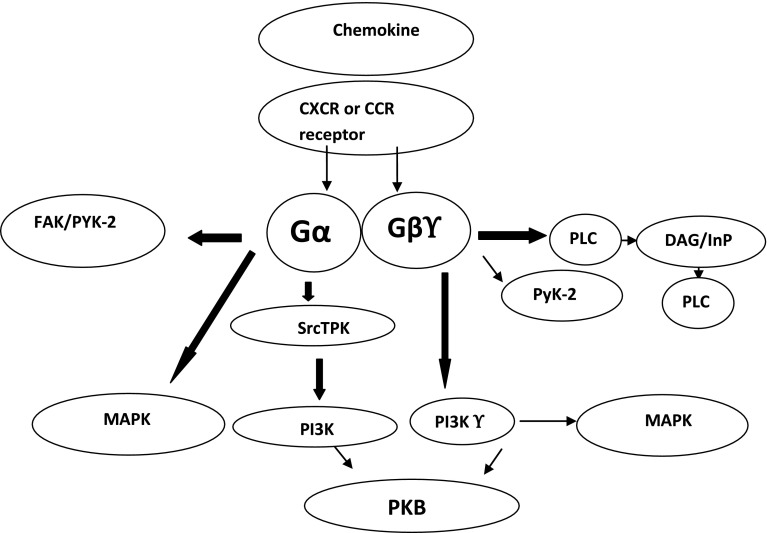
Chemokine/chemokine receptor interactions are important, which starts signaling (Fig. 1) through which wound healing, infection, and tissue maintenance occur. The expression of chemokine and chemokine receptors are often strongly up-regulated during tumorigenesis in case of different cancers like breast, lung, prostate, colon, ovary, and bladder [2]. C–C chemokine ligand 2 (CCL2), also known as monocyte chemo-attractant protein-1 (MCP-1), which is a member of the C–C beta chemokine family that is produced by macrophages, fibroblasts, and endothelial cells to stimulate chemotaxis of monocyte/macrophages and other inflammatory cells through its receptor, Chemokine receptor 2 (CCR2) [3]. Macrophage and lymphocytes infiltrating tumors are controlled by numerous cytokines, including macrophage chemoattractant protein 1 [4]. The polymorphism at A2518G in the regulatory region of the MCP-1 gene influences MCP-1 expression in response to inflammatory stimuli [5]. CCR2 gene is localized on chromosome 3p21 within a cluster of chemokine receptor genes. It has two isoforms; CCR2A and CCR2B products of the CCR2 gene as a result of alternative splicing. A single nucleotide polymorphism (SNP) of G to A at position 190 of CCR2 gene changes amino acid valine (GTC) to isoleucine (ATC) at codon 64 [6]. C–C chemokine ligand 2 and its receptor CCR2 play a key role in promoting tumorigenesis and metastasis via distinct mechanisms [7]. Inflammation involves a complex interaction of gene networks and is largely self-regulating, thus it is reasonable to assume that certain combinations of alleles may contribute to an imbalanced immune response and increased disease risk. This genetic polymorphism promises to help define pathophysiological mechanisms, to identify individuals at risk for disease, and to suggest novel targets for drug treatment. One of the main characteristics of cancer cells is their ability to proliferate, invade the surrounding tissues, and migrate to distant organs and form metastasis, thereby resulting in the emergence of disseminated metastases, which remains the primary cause of mortality in cancer patients [8–11].
Fig. 1.
Chemokine signaling (self-drawn)
The primary focus of this study is an attempt to investigate frequency distribution of CCL2I/D,CCL2 A2518G, and CCR2 V64I genes polymorphism by using a Polymerase chain reaction (PCR)-based restriction analysis in unrelated normal healthy individuals from North India thereby translating the observations in assisting high-risk screening of human exposed to environmental carcinogens and cancer predisposition in different ethnic groups., thus providing basis for future epidemiological and clinical studies.
Materials and Methods
Subjects
Healthy and genetically unrelated individuals visiting the hospital for a routine checkup or health awareness camps and hospital employees were recruited as the controls (n = 200). All the controls were age and sex matched with similar ethnicity and had no evidence of malignancy or chronic disease. The mean age of the controls was 58.5 years, and M:F ratio as 175:25. The participation rate was 100 % and blood samples were available for all subjects. Ethnicity was based on self-report and categorized as North Indian. An epidemiologic questionnaire was designed for study participants to collect data on demographic characteristics, smoking history, occupation history, and other lifestyle factors. At the end of the interview, a 5-ml blood sample was drawn into coded tubes.
Informed consent was taken from all subjects when interviewing for the demographic details and blood sample collection. The Ethical Review Board of the Institute approved the study.
We evaluated three chemokine genes, CCR2 V64I (rs1799864), CCL2 I/D (rs3917887), and CCL2 2518 A>G (rs1024611) and identified a sufficient number of epidemiologic studies on chemokine to conduct a comparative analysis for genetic polymorphisms in inflammatory genes, focusing on CCL2 and its receptor CCR2.
DNA Extraction
Five ml of blood was collected in EDTA vials and DNA was extracted from blood lymphocytes using a ‘salting out’ method [12].
Genotyping
Polymorphisms in chemokine CCL2I/D, CCL2 2518 A>G and CCR2V64I genes were analyzed using PCR–RFLP. Primer detail and PCR conditions of CCL2I/D, CCL2 2518 A>G [13] and CCR2V64I [14] respectively. Genotyping was done on 15 % PAGE using molecular weight markers and visualized after staining with ethidium bromide. Positive and negative controls were used in each genotyping assay, and 10 % of the samples were randomly selected and run in duplicates with 100 % concordance. The results were reproducible with no discrepancy in genotyping.
Prevalence of Gene Variants
We conducted a MEDLINE search using CCR2 V64I, CCL2 I/D, and CCL2 2518 A>G and ‘‘polymorphism’’ for papers published uptill 2012. The search was limited to human subjects without language restriction. For case–control studies, only genotype frequencies for the control population were considered. Studies that reported only allele frequencies and no genotype frequencies were not included. Studies based on fewer than 90 persons were excluded. When more than one article was identified for the same study population, we included the most recent publication. We identified ten publications reporting on the prevalence of CCR2 V64I polymorphism, three publications on CCL2 I/D, six studies on CCL2 2518 A>G, which were subsequently used for comparison with our study.
Statistical Analysis
Pearson’s χ2 test was done to compare the genotype and allelic frequencies of different populations using the computer software SPSS for windows (version 11.5). Court-Lab (web-based software) was used to examine Hardy–Weinberg equilibrium (www.tufts.edu). P < 0.05 was considered statistically significant.
Results
The distribution of CCR2 V64I, CCL2 I/D, and CCL2 2518 A>G genotypes and allele frequencies in northern Indian population are shown in Table 1.
Table 1.
Genotypes and allele frequency distribution of CCR2 V64I, CCL2 I/D and CCL2 2518A>G gene polymorphism in North India
| Gene | Genotype | Observed, n (%) | Expected, n (%) | Minor allele frequency | P value (HWE) |
|---|---|---|---|---|---|
| CCR2 V64I | GG | 126 (63) | 129.6 (64.8) | 19.5 | 0.251 |
| G/A | GA | 70 (35) | 62.8 (31.4) | ||
| (rs1799864) | AA | 4 (2) | 7.6 (3.8) | ||
| CCL2 I/D | II | 84 (42) | 83.2 (41.6) | 35.5 | 0.862 |
| DD | ID | 90 (45) | 91.6 (45.8) | ||
| (rs3917887) | DD | 26 (13) | 25.2 (12.6) | ||
| CCL2 2518 | AA | 81 (40.5) | 83.9 (41.9) | 35.3 | 0.532 |
| A/G | AG | 97 (48.5) | 91.3 (45.6) | ||
| (rs1024611) | GG | 22 (11.0) | 24.9 (12.4) |
Genotype distributions were in agreement with Hardy–Weinberg equilibrium at all polymorphic sites in three genes. The frequency distribution of different genotypes and alleles of these three genes with different populations with reference to ours were compared (Tables 2, 3, 4) using χ2 tests. The minor variant allele frequency in our population was as follows: 19.5 % for CCR2 V64I, 35.5 % for CCL2 I/D, 35.3 % for CCL2 2518 A>G.
Table 2.
Genotypes and allele frequency distribution of CCR2 V64I gene polymorphism in various populations and P values in comparison to Northern Indian population
| Gene | Country/ethnicity | n | Age (years), | Genotype | P | Aa | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| Mean age ± SD | GG | GA | AA | ||||||
|
CCR2 V64I (rs1799864) |
North India | 200 | 56. 8 ± 10.8 | 126 (63.0) | 70 (35.0) | 4 (2.0) | Ref | 19.5 | Present study |
| North India | 277 | 60.24 ± 8.19 | 201 (72.6) | 71 (25.6) | 5 (1.8) | 0.081 | 14.6 | Singh et al. [19] | |
| North India | 225 | >10(17.07 ± 6.69) | 189 (84.0) | 34 (15.0) | 2 (1.0) | <0.001 | 9 | Prasad et al. [20] | |
| South India | 206 | 31.3 ± 8.4 | 173 (84.0) | 30 (14.6) | 3 (1.4) | <0.001 | 8.7 | Alagarasu et al. [17] | |
| Northern Swedish | 148 | 64 (24 ± 89) | 112 (75.7) | 34 (23.0) | 2 (1.0) | 0.042 | 27.8 | Zheng et al. [21] | |
| Czech | 277 | 49.3 ± 7.9 | 218 (78.7) | 58 (20.9) | 1 (0.4) | <0.001 | 10.8 | Petrkova et al. [22] | |
| Black South African | 305 | 42.3 ± 9.0 | 189 (62.0) | 112 (37.0) | 4 (1.0) | 0.786 | 19.5 | Chatterjee et al. [16] | |
| Mixed-ancestry | 1073 | 44.3 ± 8.4 | 704 (66) | 356 (33) | 13 (1.0) | 0.569 | 17.5 | Chatterjee et al. [16] | |
| Japanese | 122 | 44.4 ± 14.1 | 66 (54.1) | 48 (39.3) | 8 (6.6) | 0.064 | 26.2 | Hizawa et al. [23] | |
| North India | 180 | 36.3 ± 12.4 | 139 (77.1) | 41 (22.8) | 0 (0.0) | 0.004 | 11.4 | Manchanda et al. [24] | |
| Turkey | 140 | 52.34 ± 11.91 | 112 (80.0) | 24 (17.9) | 4 (2.9) | 0.001 | 11.4 | Bektas-Kayhan et al. [25] | |
Bold values represents significant risk
a Minor or variant allele frequency
Table 3.
Genotypes and allele frequency distribution of CCL2 I/D gene polymorphism in various populations and P values in comparison to Northern Indian population
| Gene | Country/ethnicity | n | Age (years), | Genotype | P | Da | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| Mean age ± SD | II | ID | DD | ||||||
|
CCL2 I/D (rs3917887) |
North India | 200 | 56.8 ± 10.8 | 84 (42.0) | 90 (45) | 26 (13) | Ref | 35.5 | Present study |
| North India | 255 | 58.1 ± 8.0 | 89 (35.0) | 82 (32.0) | 84 (33.0) | <0.001 | 49 | Ahluwalia et al. [13] | |
| South India | 92 | 53.5 ± 12.1 | 29 (32) | 34 (36.0) | 29 (32) | 0.001 | 50 | Ahluwalia et al. [13] | |
| Australia | 160 | 82 (51.2) | 63 (39.4) | 15 (9.4) | 0.189 | 29.1 | Chinoy et al. [26] | ||
Bold values represents significant risk
a Minor or variant allele frequency
Table 4.
Genotypes and allele frequency distribution of CCL2 2518 A>G gene polymorphism in various populations and P values in comparison to Northern Indian population
| Gene | Country/ethnicity | n | Age (years), | Genotype | P | Ga | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| Mean age ± SD | AA | AG | GG | ||||||
| CCL2 2518 A>G (rs1024611) | North India | 200 | 56.8 ± 10.8 | 81(40.5) | 97(48.5) | 22(11.0) | Ref | 35.3 | Present study |
| North India | 255 | 58.1 ± 8.0 | 122 (48.0) | 97 (38.0) | 36 (14.0) | 0.079 | 33.0 | Ahluwalia et al. [13] | |
| South India | 206 | 31.3 ± 8.4 | 93 (45.8) | 81 (39.9) | 29 (14.3) | 0.201 | 37.4 | Alagarasu et al. [17] | |
| Germany | 140 | 38 ± 12.3 | 81 (57.9) | 50 (35.7) | 9 (6.4) | 0.006 | 24.25 | Steinmetz et al. [27] | |
| Poland | 323 | 59.7 ± 11.2 | 154 (48.0) | 145 (45.6) | 24 (7.0) | 0.169 | 30 | Kruszyna et al. [18] | |
| Poland | 325 | 60.7 ± 12.4 | 209 (64.0) | 101 (31.0) | 15 (5) | <0.001 | 20.0 | Buraczynska et al. [28] | |
| Turkey | 140 | 52.34 ± 11.91 | 94 (67.1) | 45 (32.1) | 1 (0.7) | <0.001 | 16.8 | Bektas-Kayhan et al. [25] | |
Bold values represents significant risk
a Minor or variant allele frequency
In case of CCR2 V64I significantly genotype and allele distribution were observed in two North Indian populations, South India, Northern Sweden, Czech, and Turkey population as compared to our population. Study of CCL2 I/D polymorphism is not common worldwide, only few studies have been reported. Based on reports, significantly different pattern of CCL2 I/D polymorphism and allele frequency were reported only in North India and South India. Genotype and allele distribution of CCL2 2518 A>G polymorphism was significantly different from North India, South India, Germany, Poland, and Turkey as compared to our population.
Discussion
Polymorphisms in cytokine/chemokine genes may result in inter individual variation in transcriptional regulation, and thus in differential inflammatory and pro-inflammatory molecules production.
Single nucleotide polymorphisms are scattered throughout the genome and high degree of variability make these informative genetic markers useful for disease susceptibility Functional polymorphism of chemokine is thought to be of particular importance for implication in the pathogenesis of complex genetic disorders. Due to marked differences in the distribution of chemokine gene polymorphisms between various ethnicities, the data from ‘normal healthy’ populations are of special interest for the adequate evaluation of the relevance of the investigated genetic markers in susceptibility, manifestation, prognosis or treatment of diseases. However, it is noteworthy to conduct extensive investigations about the distribution of these genes in different ethnic groups.
Indian population is believed to be most diverse because of different socio-cultural traditions. The study of genetic variation can elucidate critical determinants in environmental exposure and cancer, which could have future implications for preventive and early intervention strategies. The differences in allele frequencies detected among these studies might be due to ethnic variation, heterogeneity of study populations, and different sample sizes. The variation in our Indian population from other world population signifies the impact of ethnicity. It is well recognized that ethnic background may influence the susceptibility to suffer from certain diseases [15].
In the CCR2 V64I polymorphism, the (A*) allele frequency in Indian population was 19.5 %, which was significantly lower in two North India population, South India, Czech, Turkey and higher in Northern Swedish and but no significant difference was observed from other North India population, Black South Africa, mixed-ancestry and Japan. The (D*) allele frequency in CCL2 I/D polymorphism was 35.5 % in our population. This was significantly different and higher from North India and South India and no significant difference was observed from Australia. In CCL2 2518 A>G polymorphism, the (G*) allele frequency in Indian population was 35.3 %, which was significantly different and lower in Germany, Poland, and Turkey population while no significant difference was observed from North India, South India and other Poland population.
In a study from Black South Africa and mixed-ancestry by Chatterjee et al. 2010, the minor variant allele frequencies were found to be almost similar with our northern population for CCR2 V64I (19.5 vs. 19.5 % and 17.5 respectively) [16] whereas in case of CCL2 2518 A>G, a study from North India, South India and Poland by Ahluwalia et al. 2009, Alagarasu et al. 2009, and Kruszyna et al. 2010, the minor variant allele frequencies were found to be almost similar with our northern population i.e. (35.3 vs. 33 %, 37.4 %, and 30 % respectively) [13; 17; 18]. This suggested that there was no variability in two geographical zones from India. Large and combined analyses may be preferred to minimize the likelihood of both false-positive and false-negative results. When appropriate, confounding factors should be controlled for, with particular consideration of race and ethnicity. There are differences in the prevalence of chemokine polymorphisms across different populations. Hence, it is important to keep in mind that a susceptibility factor in one population may not hold true for another.
Such kind of study may form the basis for future establishment of epidemiological and clinical databases. This study suggest that CCL2 and its receptor CCR2 polymorphisms may be biomarkers of disease susceptibility and may be contributing factors in the risk of cancer development. A single larger study with thousands of subjects and tissue-specific biochemical and biological characterization is warranted to further evaluate potential gene-to-gene and gene-to-environment interactions on chemokine gene polymorphisms and cancer risk. The differences in these genes distribution between North Indian healthy population and other ethnic groups may help in building a profile that would help in assessing the disease predisposition and prevalence.
Acknowledgments
We are thankful to DBT (Department of Biotechnology, New Delhi) and ICMR (Indian Council of Medical Research, New Delhi) for providing financial support for the study. We are also thankful to the volunteers for providing the blood samples.
Conflict of interest
There is no conflict of interest among the authors.
References
- 1.Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006;7(12):243. doi: 10.1186/gb-2006-7-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raman D, Baugher PJ, Thu YM, Richmond A. Role of chemokines in tumor growth. Cancer Lett. 2007;256(2):137–165. doi: 10.1016/j.canlet.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazennec G, Richmond A. Chemokines and chemokine receptors: new insights into cancer-related inflammation. Trends Mol Med. 2010;16(3):133–144. doi: 10.1016/j.molmed.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22(2):231–237. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Rovin BH, Lu L, Saxena R. A novel polymorphism in the MCP-1 gene regulatory region that influences MCP-1 expression. Biochem Biophys Res Commun. 1999;259(2):344–348. doi: 10.1006/bbrc.1999.0796. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama EE, Tanaka Y, Nagai Y, Iwamoto A, Shioda T. A CCR2-V64I polymorphism affects stability of CCR2A isoform. AIDS. 2004;18(5):729–738. doi: 10.1097/00002030-200403260-00003. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Patel L, Pienta KJ. CC chemokine ligand 2 (CCL2) promotes prostate cancer tumorigenesis and metastasis. Cytokine Growth Factor Rev. 2010;21(1):41–48. doi: 10.1016/j.cytogfr.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 9.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Ye S. Polymorphism in matrix metalloproteinase gene promoters: implication in regulation of gene expression and susceptibility of various diseases. Matrix Biol. 2000;19(7):623–629. doi: 10.1016/S0945-053X(00)00102-5. [DOI] [PubMed] [Google Scholar]
- 11.Hynes RO. Metastatic potential: generic predisposition of the primary tumor or rare, metastatic variants—or both? Cell. 2003;113(7):821–823. doi: 10.1016/S0092-8674(03)00468-9. [DOI] [PubMed] [Google Scholar]
- 12.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahluwalia TS, Khullar M, Ahuja M, Kohli HS, Bhansali A, Mohan V, et al. Common variants of inflammatory cytokine genes are associated with risk of nephropathy in type 2 diabetes among Asian Indians. PLoS ONE. 2009;4(4):e5168. doi: 10.1371/journal.pone.0005168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narter KF, Agachan B, Sozen S, Cincin ZB, Isbir T. CCR2-64I is a risk factor for development of bladder cancer. Genet Mol Res. 2010;9(2):685–692. doi: 10.4238/vol9-2gmr829. [DOI] [PubMed] [Google Scholar]
- 15.Kittles RA, Weiss KM. Race, ancestry, and genes: implications for defining disease risk. Annu Rev Genomics Hum Genet. 2003;4:33–67. doi: 10.1146/annurev.genom.4.070802.110356. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee K, Dandara C, Hoffman M, Williamson AL. CCR2-V64I polymorphism is associated with increased risk of cervical cancer but not with HPV infection or pre-cancerous lesions in African women. BMC Cancer. 2010;10:278. doi: 10.1186/1471-2407-10-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alagarasu K, Selvaraj P, Swaminathan S, Raghavan S, Narendran G, Narayanan PR. CCR2, MCP-1, SDF-1a and DC-SIGN gene polymorphisms in HIV-1 infected patients with and without tuberculosis. Indian J Med Res. 2009;130(4):444–450. [PubMed] [Google Scholar]
- 18.Kruszyna L, Lianeri M, Rubis B, Knuła H, Rybczyńska M, Grodecka-Gazdecka S, et al. CCL2 -2518 A/G single nucleotide polymorphism as a risk factor for breast cancer. Mol Biol Rep. 2011;38(2):1263–1267. doi: 10.1007/s11033-010-0225-9. [DOI] [PubMed] [Google Scholar]
- 19.Singh R, Kapoor R, Srivastava A, Mittal RD. Impact of chemokine receptor CCR2 and CCR5 gene polymorphism on allograft outcome in North Indian renal transplant recipients. Scand J Immunol. 2009;69(1):51–56. doi: 10.1111/j.1365-3083.2008.02192.x. [DOI] [PubMed] [Google Scholar]
- 20.Prasad P, Tiwari AK, Kumar KM, Ammini AC, Gupta A, Gupta R, et al. Association of TGFbeta1, TNFalpha, CCR2 and CCR5 gene polymorphisms in type-2 diabetes and renal insufficiency among Asian Indians. BMC Med Genet. 2007;8:20. doi: 10.1186/1471-2350-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng B, Wiklund F, Gharizadeh B, Sadat M, Gambelunghe G, Hallmans G, et al. Genetic polymorphism of chemokine receptors CCR2 and CCR5 in Swedish cervical cancer patients. Anticancer Res. 2006;26(5B):3669–3674. [PubMed] [Google Scholar]
- 22.Petrkova J, Cermakova Z, Drabek J, Lukl J, Petrek M. CC chemokine receptor (CCR)2 polymorphism in Czech patients with myocardial infarction. Immunol Lett. 2003;88(1):53–55. doi: 10.1016/S0165-2478(03)00053-1. [DOI] [PubMed] [Google Scholar]
- 23.Hizawa N, Yamaguchi E, Furuya K, Jinushi E, Ito A, Kawakami Y. The role of the C–C chemokine receptor 2 gene polymorphism V64I (CCR2-64I) in sarcoidosis in a Japanese population. Am J Respir Crit Care Med. 1999;159(6):2021–2023. doi: 10.1164/ajrccm.159.6.9810020. [DOI] [PubMed] [Google Scholar]
- 24.Manchanda PK, Singh R, Mittal RD. Cytokine (IL-10 -1082 and -819) and chemokine receptor (CCR2 and CCR5) gene polymorphism in North Indian patients with end-stage renal disease. DNA Cell Biol. 2009;28(4):177–183. doi: 10.1089/dna.2008.0822. [DOI] [PubMed] [Google Scholar]
- 25.Bektas-Kayhan K, Unur M, Boy-Metin Z, Cakmakoglu B. MCP-1 and CCR2 gene variants in oral squamous cell carcinoma. Oral Dis. 2012;18(1):55–59. doi: 10.1111/j.1601-0825.2011.01843.x. [DOI] [PubMed] [Google Scholar]
- 26.Chinoy H, Salway F, Fertig N, Tait BD, Oddis CV, Ollier WE, et al. Monocyte chemotactic protein-1 single nucleotide polymorphisms do not confer susceptibility for the development of adult onset polymyositis/dermatomyositis in UK Caucasians. Rheumatology. 2007;46(4):604–607. doi: 10.1093/rheumatology/kel359. [DOI] [PubMed] [Google Scholar]
- 27.Steinmetz OM, Panzer U, Harendza S, Mertens PR, Ostendorf T, Floege J, et al. No association of the -2518 MCP-1 A/G promoter polymorphism with incidence and clinical course of IgA nephropathy. Nephrol Dial Transplant. 2004;19(3):596–601. doi: 10.1093/ndt/gfg577. [DOI] [PubMed] [Google Scholar]
- 28.Buraczynska M, Bednarek-Skublewska A, Buraczynska K, Ksiazek A. Monocyte chemoattractant protein-1 (MCP-1) gene polymorphism as a potential risk factor for cardiovascular disease in hemodialyzed patients. Cytokine. 2008;44(3):361–365. doi: 10.1016/j.cyto.2008.10.001. [DOI] [PubMed] [Google Scholar]