Table 2. Data collection from a bovine trypsin crystal at 13.2keV in several orientations and SHELXD/E results.
Values in parentheses are for the highest resolution shell (1.91.8).
| Alignment | (0 0 1) | (1 3 1) | (0 1 1) | (2 1 1) |
|---|---|---|---|---|
| Position of data collection | 1 | 2 | 2 | 1 |
| Space group | P212121 | |||
| Unit-cell parameters () | a = 54.3, b = 58.1, c = 66.8 | |||
| No. of images | 360 | 360 | 360 | 360 |
| () | 151.1 | 148.0 | 50.6 | 109.7 |
| () | 109.7 | 146.7 | 230.4 | 22.1 |
| Unique reflections | 24991 (3560) | 25630 (3697) | 25614 (3689) | 25632 (3698) |
| Completeness (%) | 98.1 (97.3) | 100.0 (100.0) | 99.9 (99.9) | 99.9 (100.0) |
| Multiplicity† | 14.5 [7.7] | 14.2 [7.5] | 14.2 [7.6] | 14.2 [7.5] |
| I/(I) | 74.8 (35.9) | 57.4 (24.4) | 65.2 (31.4) | 60.0 (31.2) |
| R meas ‡ (%) | 2.7 (7.2) | 3.5 (9.4) | 3.1 (7.8) | 3.5 (7.6) |
| R p.i.m. § (%) | 0.7 (1.9) | 0.9 (2.5) | 0.8 (2.0) | 0.9 (2.0) |
| R anom ¶ (%) | 0.8 | 0.9 | 0.8 | 0.9 |
| Anomalous signal to noise†† | 0.94 (0.84) | 0.85 (0.79) | 0.85 (0.78) | 0.83 (0.76) |
| SHELXD | ||||
| FIND/DSUL | 14/6 | 14/6 | 14/6 | 14/6 |
| Resolution for SHELXD ‡‡ () | 2.3 | 2.3 | 2.3 | 2.3 |
| CCall | 22.8 | 16.0 | 16.0 | 19.0 |
| CCweak | 11.0 | 4.4 | 5.1 | 9.3 |
| PATFOM | 3.3 | 2.6 | 2.9 | 3.1 |
| SHELXE | ||||
| Solvent content (%) | 57 | 57 | 57 | 57 |
| Contrast | 0.52 | 0.49 | 0.5 | 0.59 |
| Connectivity | 0.78 | 0.71 | 0.71 | 0.73 |
| Pseudo-free CC (%) | 72 | 55.3 | 53.75 | 58.8 |
Multiplicity of the native and anomalous (in square brackets) data.
R
meas = 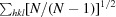
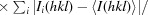
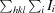 (hkl) is the multiplicity (N) independent R
merge.
(hkl) is the multiplicity (N) independent R
merge.
R
p.i.m. = 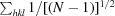
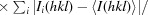
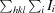 (hkl) is a precision-indicating R factor.
(hkl) is a precision-indicating R factor.
R
anom = 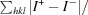
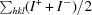 is the ratio of the mean anomalous intensity difference to the mean reflection intensity.
is the ratio of the mean anomalous intensity difference to the mean reflection intensity.
Anomalous signal to noise = 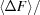
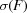 is the ratio of the anomalous difference to the noise.
is the ratio of the anomalous difference to the noise.
Resolution used for heavy-atom search.
