Table 3. Data collection and SHELXD and SOLOMON results for an SeMet-substituted domain from a protein involved in piRNA biogenesis collected in two orientations.
Values in parentheses are for the highest resolution shell (2.952.80).
| Alignment | (0 0 1) | None | Combined |
|---|---|---|---|
| Space group | P212121 | ||
| Unit-cell parameters () | a = 35.2, b = 100.8, c = 146.3 | ||
| No. of images | 100 | 100 | 100 + 100 |
| () | 52.0 | 0 | |
| () | 150.01 | 0 | |
| Unique reflections | 13160 (1873) | 13576 (1963) | 13594 (1948) |
| Completeness (%) | 97.8 (96.9) | 99.8 (99.5) | 100.0 (99.9) |
| Multiplicity† | 4.0 [2.2] | 3.9 [2.1] | 7.7 [4.1] |
| I/(I) | 12.5 (2.4) | 10.3 (1.8) | 13.7 (2.9) |
| R meas ‡ (%) | 14.5 (78.6) | 16.7 (106.8) | 16.6 (72.7) |
| R p.i.m. § (%) | 7.1 (38.0) | 8.4 (52.8) | 5.9 (32.6) |
| R anom (%) | 10.4 | 11.7 | 9.4 |
| Wilson B factor (2) | 62.7 | 67.2 | 60.7 |
| Anomalous signal to noise | 1.35 (0.72) | 1.16 (0.68) | 1.45 (0.75) |
| SHELXD | |||
| FIND | 20 | 20 | 20 |
| Resolution for SHELXD () | 3.3 | 3.3 | 3.3 |
| CCall | 47.9 | 33.2 | 50.8 |
| CCweak | 18.8 | 7.1 | 18.9 |
| PATFOM | 5.2 | 3.8 | 4.9 |
| SOLOMON † | |||
| Overall correlation on |E|2 | 0.41 versus 0.34 | 0.44 versus 0.35 | |
Multiplicity of the native and anomalous (in square brackets) data.
R
meas = 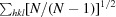
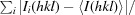
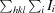 (hkl) is the multiplicity (N) independent R
merge.
(hkl) is the multiplicity (N) independent R
merge.
R
p.i.m. = 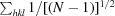
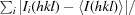
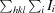 (hkl) is a precision-indicating R factor.
(hkl) is a precision-indicating R factor.
R
anom = 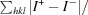
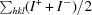 is the ratio of the mean anomalous intensity difference to the mean reflection intensity.
is the ratio of the mean anomalous intensity difference to the mean reflection intensity.
Anomalous signal to noise = 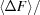
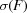 is the ratio of the anomalous difference to the noise.
is the ratio of the anomalous difference to the noise.
Resolution used for the heavy-atom search.
The correlation coefficient calculated between the structure-factor amplitudes of the ‘observed’ data and the modified map using SOLOMON (Abrahams Leslie, 1996 ▶) for the correct versus the incorrect hand.
