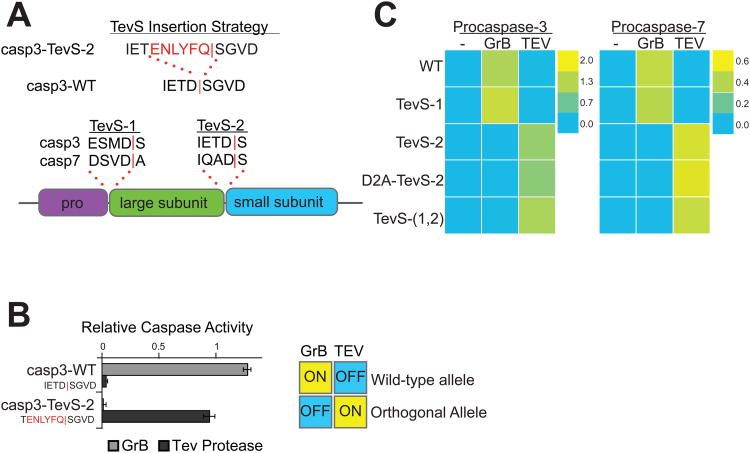
Figure 2. Design and In Vitro Characterization of Orthogonal Procaspase-3 and -7 Alleles.
(A) We employed the indicated TevS insertion strategy to convert caspase sites into SNIPer-cleavage sites as shown for caspase-3. The sequences (P4-P1′ with a red bar to denote the scissile bond) and location of site-1 and site-2 are shown in the context of executioner caspase domain structure below. (B) In vitro activation of wild-type procaspase-3 (Casp3-WT) or Casp3-TevS was assayed by expressing each allele in E. coli, treating with either rat Granzyme-B (20nM) or TEV Protease(1.1uM) for 1h, and measuring caspase activity with 10uM Ac-DEVD-AFC. Data are presented as the average rate of substrate cleavage (RFU s-1) from an experiment performed in triplicate (Error Bars±SD). (C) Caspase activity of the procaspase-3 and -7 matrix of orthogonal alleles was performed as above and is presented as a heat map. Abbreviations: WT, wild-type; TevS-1, TevS insertion at site-1; TevS-2, TevS insertion at site-2; D2A-TevS-2, TevS insertion at site-2 with a non-cleavable site-1; and TevS-(1,2), TevS insertions at site-1 and site-2. See also Figure S2.